3IH0
 
 | | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP | | Descriptor: | GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized sugar kinase PH1459 | | Authors: | Kumar, G, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-29 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an uncharacterized sugar kinase PH1459 from Pyrococcus horikoshii in complex with AMP-PNP
To be Published
|
|
1J27
 
 | | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7A resolution | | Descriptor: | hypothetical protein TT1725 | | Authors: | Seto, A, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-26 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a hypothetical protein, TT1725, from Thermus thermophilus HB8 at 1.7 A resolution
Proteins, 53, 2003
|
|
1J2W
 
 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3I7V
 
 | | Crystal structure of AP4A hydrolase complexed with AP4A (ATP) (aq_158) from Aquifex aeolicus Vf5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DNO
 
 | | Molecular structure for the HIV-1 gp120 trimer in the CD4-bound state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3I7U
 
 | | Crystal structure of AP4A hydrolase (aq_158) from Aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3CNC
 
 | |
2NYG
 
 | |
1RTX
 
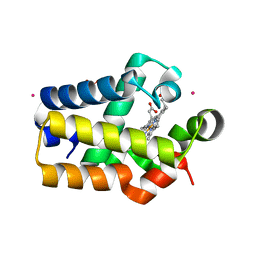 | | Crystal Structure of Synechocystis Hemoglobin with a Covalent Heme Linkage | | Descriptor: | CADMIUM ION, Cyanoglobin, POTASSIUM ION, ... | | Authors: | Hoy, J.A, Kundu, S, Trent, J.T, Ramaswamy, S, Hargrove, M.S. | | Deposit date: | 2003-12-10 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of synechocystis hemoglobin with a covalent heme linkage.
J.Biol.Chem., 279, 2004
|
|
3HPD
 
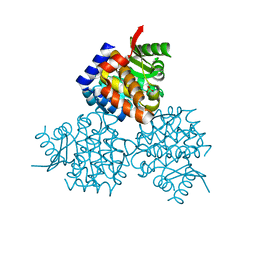 | | Structure of hydroxyethylthiazole kinase protein from pyrococcus horikoshii OT3 | | Descriptor: | Hydroxyethylthiazole kinase, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Thamotharan, S, Kuramitsu, S, Yokoyama, S, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Hydroxyethylthiazole Kinase Protein from Pyrococcus Horikoshii Ot3
To be Published
|
|
2NXO
 
 | |
1O7H
 
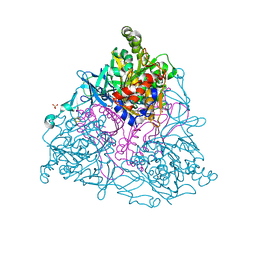 | | NAPHTHALENE 1,2-DIOXYGENASE WITH OXIDIZED RIESKE IRON SULPHUR CENTER SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-05 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7M
 
 | | NAPHTHALENE 1,2-DIOXYGENASE, BINARY COMPLEX WITH DIOXYGEN | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-11 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7G
 
 | | NAPHTHALENE 1,2-DIOXYGENASE WITH NAPHTHALENE BOUND IN THE ACTIVE SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-05 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
1O7W
 
 | | NAPHTHALENE 1,2-DIOXYGENASE, FULLY REDUCED FORM | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-11-14 | | Release date: | 2003-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Naphthalene Dioxygenase: Side-on Binding of Dioxygen to Iron
Science, 299, 2003
|
|
2BVB
 
 | | The C-terminal domain from Micronemal Protein 1 (MIC1) from Toxoplasma Gondii | | Descriptor: | MICRONEMAL PROTEIN 1 | | Authors: | Saouros, S, Edwards-Jones, B, Reiss, M, Sawmynaden, K, Cota, E, Simpson, P, Dowse, T.J, Jakle, U, Ramboarina, S, Shivarattan, T, Matthews, S, Soldati-Favre, D. | | Deposit date: | 2005-06-23 | | Release date: | 2005-10-12 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | A Novel Galectin-Like Domain from Toxoplasma Gondll Micronemal Protein 1 Assists the Folding, Assembly,and Transport of a Cell-Adhesion Complex.
J.Biol.Chem., 280, 2005
|
|
3HP0
 
 | |
1KEB
 
 | | Crystal Structure of Double Mutant M37L,P40S E.coli Thioredoxin | | Descriptor: | COPPER (II) ION, Thioredoxin 1 | | Authors: | Rudresh, Jain, R, Dani, V, Mitra, A, Srivastava, S, Sarma, S.P, Varadarajan, R, Ramakumar, S. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Consequences of Replacement of an alpha-helical Pro Residue in E.coli Thioredoxin
PROTEIN ENG., 15, 2002
|
|
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
3DNN
 
 | | Molecular structure for the HIV-1 gp120 trimer in the unliganded state | | Descriptor: | HIV-1 envelope glycoprotein gp120 | | Authors: | Borgnia, M.J, Liu, J, Bartesaghi, A, Sapiro, G, Subramaniam, S. | | Deposit date: | 2008-07-02 | | Release date: | 2008-08-19 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Molecular architecture of native HIV-1 gp120 trimers.
Nature, 455, 2008
|
|
3DZU
 
 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with BVT.13, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-[(2,4-DICHLOROBENZOYL)AMINO]-5-(PYRIMIDIN-2-YLOXY)BENZOIC ACID, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3E00
 
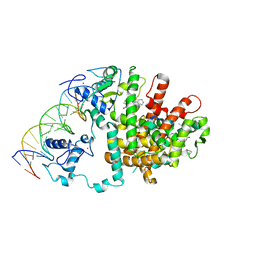 | | Intact PPAR gamma - RXR alpha Nuclear Receptor Complex on DNA bound with GW9662, 9-cis Retinoic Acid and NCOA2 Peptide | | Descriptor: | (9cis)-retinoic acid, 2-chloro-5-nitro-N-phenylbenzamide, DNA (5'-D(*DCP*DAP*DAP*DAP*DCP*DTP*DAP*DGP*DGP*DTP*DCP*DAP*DAP*DAP*DGP*DGP*DTP*DCP*DAP*DG)-3'), ... | | Authors: | Chandra, V, Huang, P, Hamuro, Y, Raghuram, S, Wang, Y, Burris, T.P, Rastinejad, F. | | Deposit date: | 2008-07-30 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA.
Nature, 456, 2008
|
|
3IXQ
 
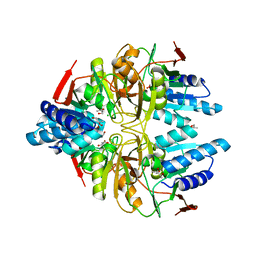 | | Structure of ribose 5-phosphate isomerase a from methanocaldococcus jannaschii | | Descriptor: | ACETATE ION, CHLORIDE ION, Ribose-5-phosphate isomerase A, ... | | Authors: | Antonyuk, S.V, Ellis, M.J, Strange, R.W, Hasnain, S.S, Bessho, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of an archaeal ribose-5-phosphate isomerase from Methanocaldococcus jannaschii (MJ1603).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3OPF
 
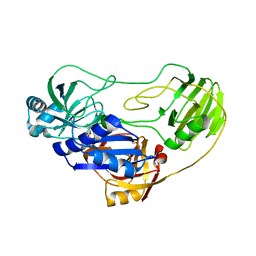 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1PVX
 
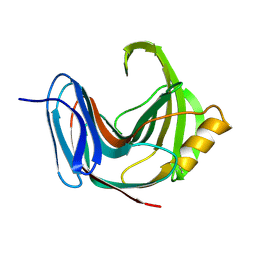 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
