7L8L
 
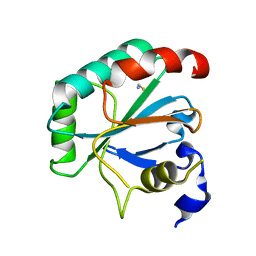 | | Crystal structure of human R152H GPX4-U46C | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8M
 
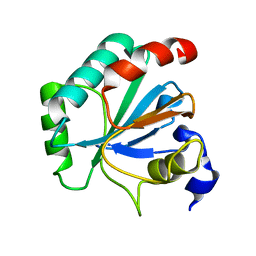 | | Crystal structure of human GPX4-U46C mutant K48L | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | Deposit date: | 2020-12-31 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
6EBM
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, transmembrane domain of subunit alpha | | Descriptor: | Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
8IYM
 
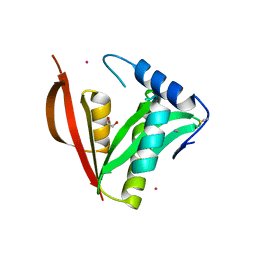 | | Crystal structure of a protein acetyltransferase, HP0935 | | Descriptor: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
8IYO
 
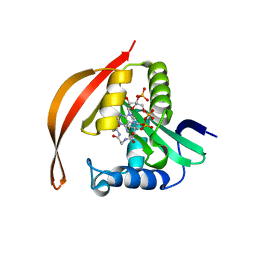 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | Descriptor: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | Authors: | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
6EBK
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EBL
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
2Q3N
 
 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
3BE3
 
 | |
1QV7
 
 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,3-DIFLUOROBENZYL ALCOHOL | | Descriptor: | 2,3-DIFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-27 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
2A4K
 
 | | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137 | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase, UNKNOWN ATOM OR ION | | Authors: | Zhou, W, Ebihara, A, Tempel, W, Yokoyama, S, Chen, L, Kuramitsu, S, Nguyen, J, Chang, S.-H, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137
To be Published
|
|
8UNH
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2POF
 
 | |
2PLR
 
 | | Crystal structure of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Rafi, Z.A, Sekar, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of dTMP kinase (st1543) from Sulfolobus Tokodaii Strain7
To be Published
|
|
2PPY
 
 | | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-01 | | Release date: | 2008-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426
To be Published
|
|
2PKP
 
 | | Crystal structure of 3-isopropylmalate dehydratase (leuD)from Methhanocaldococcus Jannaschii DSM2661 (MJ1271) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Homoaconitase small subunit, ZINC ION | | Authors: | Jeyakanthan, J, Gayathri, D.R, Velmurugan, D, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-18 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity determinants of the methanogen homoaconitase enzyme: structure and function of the small subunit
Biochemistry, 49, 2010
|
|
2PG0
 
 | | Crystal structure of acyl-CoA dehydrogenase from Geobacillus kaustophilus | | Descriptor: | Acyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Li, Y, Fu, Z.-Q, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acyl-CoA dehydrogenase from G. kaustophilus
To be Published
|
|
2PW6
 
 | | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12 | | Descriptor: | Uncharacterized protein ygiD, ZINC ION | | Authors: | Newton, M.G, Takagi, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokayama, S, Li, Y, Chen, L, Zhu, J, Ruble, J, Liu, Z.J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12.
To be Published
|
|
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
1R0R
 
 | | 1.1 Angstrom Resolution Structure of the Complex Between the Protein Inhibitor, OMTKY3, and the Serine Protease, Subtilisin Carlsberg | | Descriptor: | CALCIUM ION, Ovomucoid, subtilisin carlsberg | | Authors: | Horn, J.R, Ramaswamy, S, Murphy, K.P. | | Deposit date: | 2003-09-22 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and energetics of protein-protein interactions: the role of conformational heterogeneity in OMTKY3 binding to serine proteases
J.Mol.Biol., 331, 2003
|
|
2QQ4
 
 | | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8 | | Descriptor: | Iron-sulfur cluster biosynthesis protein IscU, ZINC ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-07-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Iron-sulfur cluster biosynthesis protein IscU (TTHA1736) from thermus thermophilus HB8
To be Published
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R26
 
 | | Crystal structure of thioredoxin from Trypanosoma brucei brucei | | Descriptor: | Thioredoxin | | Authors: | Friemann, R, Schmidt, H, Ramaswamy, S, Forstner, M, Krauth-Siegel, R.L, Eklund, H. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of thioredoxin from Trypanosoma brucei brucei
FEBS Lett., 554, 2003
|
|
