7KD8
 
 | | TtgR C137I I141W M167L F168Y mutant in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-10-08 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1A
 
 | | TtgR quadruple mutant (C137I I141W M167L F168Y) | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7K1C
 
 | | TtgR in complex with resveratrol | | 分子名称: | HTH-type transcriptional regulator TtgR, MAGNESIUM ION, RESVERATROL | | 著者 | Bingman, C.A, Nishikawa, K.K, Smith, R.W, Raman, S. | | 登録日 | 2020-09-07 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Epistasis shapes the fitness landscape of an allosteric specificity switch.
Nat Commun, 12, 2021
|
|
7KIY
 
 | | Plasmodium falciparum RhopH complex in soluble form | | 分子名称: | Cytoadherence linked asexual protein 3, High molecular weight rhoptry protein 3, High molecular weight rhoptry protein-2 | | 著者 | Schureck, M.A, Darling, J.E, Merk, A, Subramaniam, S, Desai, S.A. | | 登録日 | 2020-10-25 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake.
Elife, 10, 2021
|
|
6PLD
 
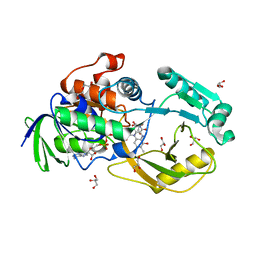 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with 6-OH-FAD - Green fraction | | 分子名称: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | 著者 | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | 登録日 | 2019-06-30 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
8IYM
 
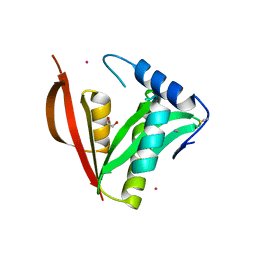 | | Crystal structure of a protein acetyltransferase, HP0935 | | 分子名称: | 1,2-ETHANEDIOL, N-acetyltransferase domain-containing protein, POTASSIUM ION, ... | | 著者 | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | 登録日 | 2023-04-05 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a protein acetyltransferase, HP0935
To be published
|
|
8IYO
 
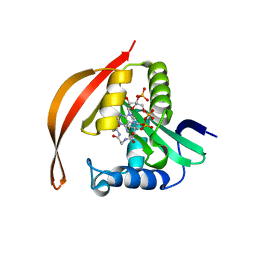 | | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form | | 分子名称: | ACETYL COENZYME *A, N-acetyltransferase domain-containing protein | | 著者 | Dadireddy, V, Mahanta, P, Kumar, A, Desirazu, R.N, Ramakumar, S. | | 登録日 | 2023-04-05 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a protein acetyltransferase, HP0935, acetyl-CoA bound form
To be published
|
|
7L8K
 
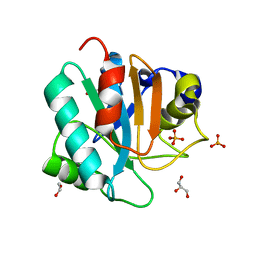 | | Crystal structure of human GPX4-U46C | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8Q
 
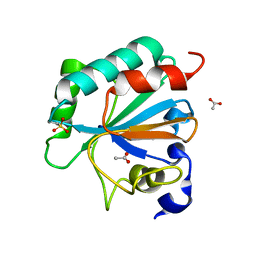 | | Crystal structure of human GPX4-U46C with oxidized Cys-46 | | 分子名称: | ACETATE ION, Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8R
 
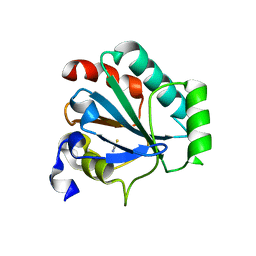 | | Crystal structure of human GPX4-U46C mutant K48A | | 分子名称: | Isoform Cytoplasmic of Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8L
 
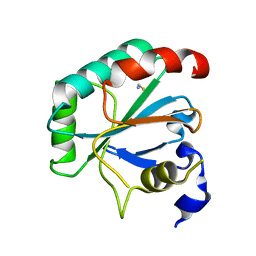 | | Crystal structure of human R152H GPX4-U46C | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Wigby, K, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
7L8M
 
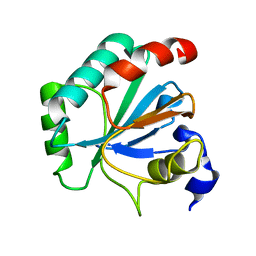 | | Crystal structure of human GPX4-U46C mutant K48L | | 分子名称: | Phospholipid hydroperoxide glutathione peroxidase | | 著者 | Forouhar, F, Liu, H, Seibt, T, Saneto, R, Friedman, J, Xia, X, Shchepinov, M.S, Ramesh, S, Conrad, M, Stockwell, B.R. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Patient-derived variant of GPX4 reveals the structural basis for its catalytic activity and degradation mechanism
Nat.Chem.Biol., 2021
|
|
6P9D
 
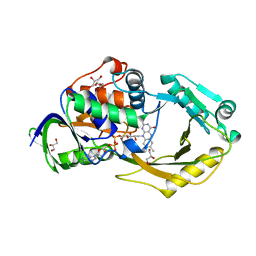 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | 著者 | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | 登録日 | 2019-06-10 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.329 Å) | | 主引用文献 | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
4YMG
 
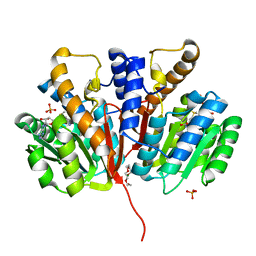 | | Crystal structure of SAM-bound Podospora anserina methyltransferase PaMTH1 | | 分子名称: | MAGNESIUM ION, PHOSPHATE ION, Putative SAM-dependent O-methyltranferase, ... | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.899 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
4YMH
 
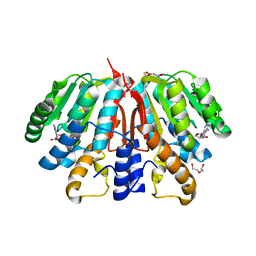 | | Crystal structure of SAH-bound Podospora anserina methyltransferase PaMTH1 | | 分子名称: | DI(HYDROXYETHYL)ETHER, Putative SAM-dependent O-methyltranferase, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Kudlinzki, D, Linhard, V.L, Chatterjee, D, Saxena, K, Sreeramulu, S, Schwalbe, H. | | 登録日 | 2015-03-06 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.876 Å) | | 主引用文献 | Structure and Biophysical Characterization of the S-Adenosylmethionine-dependent O-Methyltransferase PaMTH1, a Putative Enzyme Accumulating during Senescence of Podospora anserina.
J.Biol.Chem., 290, 2015
|
|
7MDH
 
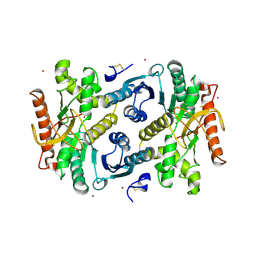 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | 分子名称: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | 著者 | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | 登録日 | 1999-02-16 | | 公開日 | 1999-06-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
7JSN
 
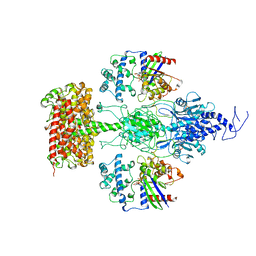 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | 分子名称: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | 登録日 | 2020-08-15 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
7XFA
 
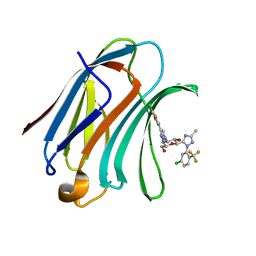 | | Structure of human Galectin-3 CRD in complex with monosaccharide inhibitor | | 分子名称: | (2~{S},3~{R},4~{R},5~{R},6~{R})-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-[2-[5-chloranyl-2-(trifluoromethyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,5-diol, Galectin-3 | | 著者 | Shukla, J, Raman, S, Ghosh, K. | | 登録日 | 2022-04-01 | | 公開日 | 2022-10-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Identification of Monosaccharide Derivatives as Potent, Selective, and Orally Bioavailable Inhibitors of Human and Mouse Galectin-3.
J.Med.Chem., 65, 2022
|
|
6EBK
 
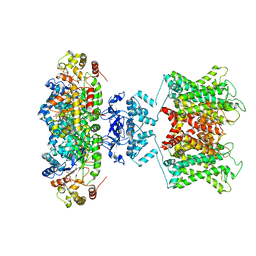 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EBL
 
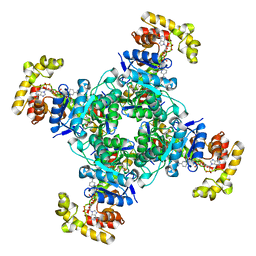 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6PWF
 
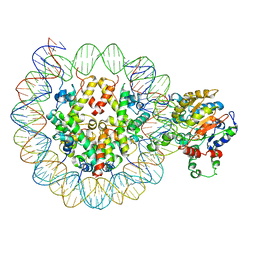 | |
6PWE
 
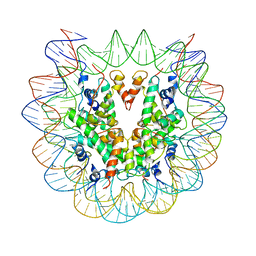 | |
6FKX
 
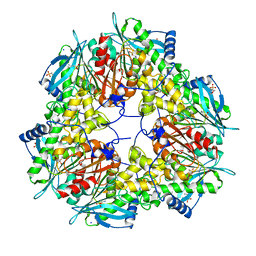 | | Crystal structure of an acetyl xylan esterase from a desert metagenome | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl xylan esterase, FORMIC ACID, ... | | 著者 | Adesioye, F.A, Makhalanyane, T.P, Vikram, S, Sewell, B.T, Schubert, W, Cowan, D.A. | | 登録日 | 2018-01-24 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Appl. Environ. Microbiol., 84, 2018
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | 著者 | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | 登録日 | 2018-03-05 | | 公開日 | 2018-06-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
7SY4
 
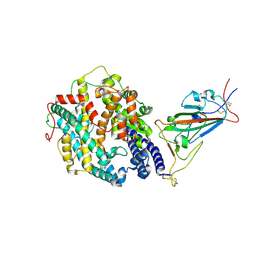 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | 登録日 | 2021-11-24 | | 公開日 | 2021-12-29 | | 最終更新日 | 2022-01-05 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
