2HZT
 
 | | Crystal Structure of a putative HTH-type transcriptional regulator ytcD | | Descriptor: | Putative HTH-type transcriptional regulator ytcD | | Authors: | Madegowda, M, Eswaramoorthy, S, Desigan, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-29 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a putative HTH-type transcription regulator ytcD
To be Published
|
|
3B59
 
 | |
2F1R
 
 | | Crystal Structure of molybdopterin-guanine biosynthesis protein B (mobB) | | Descriptor: | CHLORIDE ION, PRASEODYMIUM ION, molybdopterin-guanine dinucleotide biosynthesis protein B (mobB) | | Authors: | Damodharan, L, Eswaramoorthy, S, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of molybdopterin-guanine dinucleotide biosynthesis protein B (mobB)
To be Published
|
|
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2GS9
 
 | | Crystal structure of TT1324 from Thermus thermophilis HB8 | | Descriptor: | FORMIC ACID, Hypothetical protein TT1324, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kamitori, S, Abe, A, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-25 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of TT1324 from Thermus thermophilis HB8
To be Published
|
|
2ETF
 
 | |
2EUI
 
 | |
1IZ9
 
 | | Crystal Structure of Malate Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | MALATE DEHYDROGENASE | | Authors: | Hirose, R, Hasegawa, T, Yamano, A, Kuramitsu, S, Hamada, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Malate Dehydrogenase from Thermus themrophilus HB8
To be published
|
|
1EPW
 
 | |
3GPM
 
 | | Structure of the trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2LUO
 
 | | NMR solution structure of apo-MptpA | | Descriptor: | LOW MOLECULAR WEIGHT PROTEIN-TYROSINE-PHOSPHATASE A | | Authors: | Stehle, T, Sreeramulu, S, Loehr, F, Richter, C, Saxena, K, Jonker, H.R.A, Schwalbe, H. | | Deposit date: | 2012-06-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Apo-structure of the Low Molecular Weight Protein-tyrosine Phosphatase A (MptpA) from Mycobacterium tuberculosis Allows for Better Target-specific Drug Development.
J.Biol.Chem., 287, 2012
|
|
3HDI
 
 | | Crystal structure of Bacillus halodurans metallo peptidase | | Descriptor: | COBALT (II) ION, Processing protease, SULFATE ION, ... | | Authors: | Aleshin, A, Gramatikova, S, Strongin, A.Y, Stec, B, Liddington, R.C, Smith, J.W. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal and solution structures of a prokaryotic M16B peptidase: an open and shut case.
Structure, 17, 2009
|
|
1F1M
 
 | | CRYSTAL STRUCTURE OF OUTER SURFACE PROTEIN C (OSPC) | | Descriptor: | OUTER SURFACE PROTEIN C, ZINC ION | | Authors: | Kumaran, D, Eswaramoorthy, S, Dunn, J.J, Swaminathan, S. | | Deposit date: | 2000-05-19 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.
EMBO J., 20, 2001
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1RXF
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Schofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | Descriptor: | 32.5 KDA PROTEIN YLR351C | | Authors: | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-06-29 | | Release date: | 2001-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
1RXG
 
 | | DEACETOXYCEPHALOSPORIN C SYNTHASE COMPLEXED WITH FE(II) AND 2-OXOGLUTARATE | | Descriptor: | 2-OXOGLUTARIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION, ... | | Authors: | Valegard, K, Terwisscha Van Scheltinga, A.C, Lloyd, M.D, Hara, T, Ramaswamy, S, Perrakis, A, Thompson, A, Lee, H.J, Baldwin, J.E, Shofield, C.J, Hajdu, J, Andersson, I. | | Deposit date: | 1998-06-05 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a cephalosporin synthase.
Nature, 394, 1998
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1KRH
 
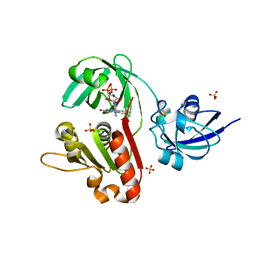 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
1DJ7
 
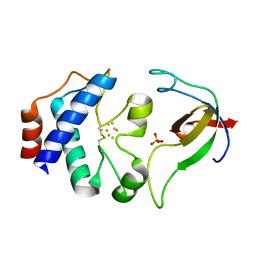 | | CRYSTAL STRUCTURE OF FERREDOXIN THIOREDOXIN REDUCTASE | | Descriptor: | FERREDOXIN THIOREDOXIN REDUCTASE: CATALYTIC CHAIN, FERREDOXIN THIOREDOXIN REDUCTASE: VARIABLE CHAIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dai, S, Schwendtmayer, C, Schurmann, P, Ramaswamy, S, Eklund, H. | | Deposit date: | 1999-12-02 | | Release date: | 2000-02-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox signaling in chloroplasts: cleavage of disulfides by an iron-sulfur cluster.
Science, 287, 2000
|
|
2NOT
 
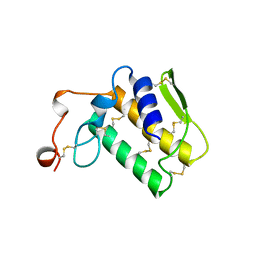 | | NOTECHIS II-5, NEUROTOXIC PHOSPHOLIPASE A2 FROM NOTECHIS SCUTATUS SCUTATUS | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Carredano, E, Westerlund, B, Persson, B, Saarinen, M, Ramaswamy, S, Eaker, D, Eklund, H. | | Deposit date: | 1997-03-03 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structures of two toxins from snake venom throw light on the anticoagulant and neurotoxic sites of phospholipase A2.
Toxicon, 36, 1998
|
|
1UUV
 
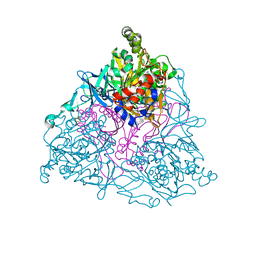 | | NAPHTHALENE 1,2-DIOXYGENASE WITH NITRIC OXIDE AND INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Karlsson, A, Parales, J.V, Parales, R.E, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2004-01-11 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | No Binding to Naphthalene Dioxygenase.
J.Biol.Inorg.Chem., 10, 2005
|
|
