2ZKD
 
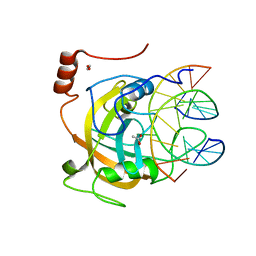 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
5C2Z
 
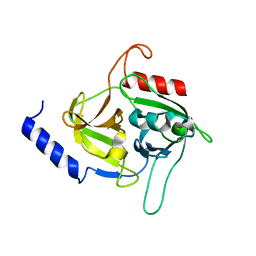 | | Molecular insights into the specificity of exfoliative toxins from Staphylococcus aureus | | Descriptor: | Exfoliative toxin D2 | | Authors: | Mariutti, R.B, Souza, T.A.C.B, Ullah, A, Zanphorlin, L.M, Murakami, M.T, Arni, R.K. | | Deposit date: | 2015-06-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9553 Å) | | Cite: | Crystal structure of Staphylococcus aureus exfoliative toxin D-like protein: Structural basis for the high specificity of exfoliative toxins.
Biochem.Biophys.Res.Commun., 467, 2015
|
|
3VYB
 
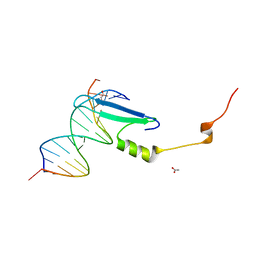 | | Crystal structure of methyl CpG binding domain of MBD4 in complex with the 5mCG/hmCG sequence | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*GP*TP*CP*(5HC)P*GP*GP*TP*AP*GP*TP*GP*AP*CP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
3VYQ
 
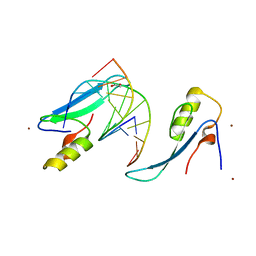 | | Crystal structure of the methyl CpG Binding Domain of MBD4 in complex with the 5mCG/TG sequence in space group P1 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*AP*TP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*GP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Otani, J, Arita, K, Kato, T, Kinoshita, M, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4
J.Biol.Chem., 288, 2013
|
|
2RR9
 
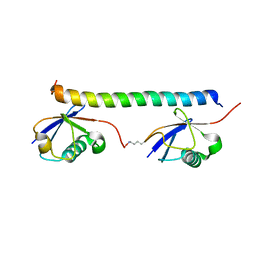 | | The solution structure of the K63-Ub2:tUIMs complex | | Descriptor: | Putative uncharacterized protein UIMC1, ubiquitin | | Authors: | Sekiyama, N, Jee, J, Isogai, S, Akagi, K, Huang, T, Ariyoshi, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2010-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the K63-Ub2:tUIMs complex
To be Published
|
|
3WO2
 
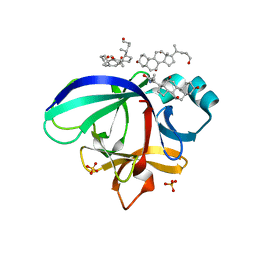 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO4
 
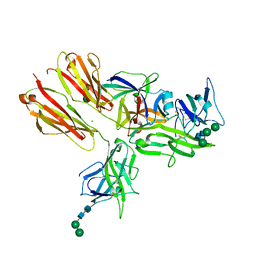 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
4DAR
 
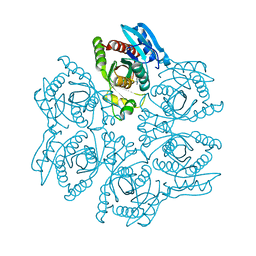 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with tubercidin | | Descriptor: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA7
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with aciclovir | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAE
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 6-chloroguanosine | | Descriptor: | 6-chloro-9-(beta-D-ribofuranosyl)-9H-purin-2-amine, ACETATE ION, CHLORIDE ION, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAN
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA6
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with ganciclovir | | Descriptor: | 9-(1,3-DIHYDROXY-PROPOXYMETHANE)GUANINE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAO
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with adenine | | Descriptor: | ADENINE, GLYCEROL, Purine nucleoside phosphorylase deoD-type | | Authors: | Giuseppe, P.O, Martins, N.H, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DA8
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 8-bromoguanosine | | Descriptor: | 8-bromoguanosine, Purine nucleoside phosphorylase deoD-type | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4DAB
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with hypoxanthine | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
5HOS
 
 | |
2ZKE
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
3BCQ
 
 | | Crystal structure of oxy-hemoglobin from Brycon cephalus | | Descriptor: | Alpha-chain hemoglobin, Beta-chain hemoglobin, OXYGEN MOLECULE, ... | | Authors: | Poy, C.D, Leopoldino, A.M, Rahal, P, de Azevedo, W.F, Rodriguez, G.O.B, Murakami, M.T. | | Deposit date: | 2007-11-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxy-hemoglobin from Brycon cephalus
To be Published
|
|
5HPC
 
 | |
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
3S2C
 
 | | Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 | | Descriptor: | Alpha-N-arabinofuranosidase | | Authors: | Souza, T.A.C.B, Santos, C.R, Souza, A.R, Oldiges, D.P, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a novel thermostable GH51 Alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1
Protein Sci., 20, 2011
|
|
1IRG
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
2ZKG
 
 | | Crystal structure of unliganded SRA domain of mouse Np95 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
1IRF
 
 | | INTERFERON REGULATORY FACTOR-2 DNA BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | INTERFERON REGULATORY FACTOR-2 | | Authors: | Furui, J, Uegaki, K, Yamazaki, T, Shirakawa, M, Swindells, M.B, Harada, H, Taniguchi, T, Kyogoku, Y. | | Deposit date: | 1997-11-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IRF-2 DNA-binding domain: a novel subgroup of the winged helix-turn-helix family.
Structure, 6, 1998
|
|
2LMK
 
 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
