1JEM
 
 | | NMR STRUCTURE OF HISTIDINE PHOSPHORYLATED FORM OF THE PHOSPHOCARRIER HISTIDINE CONTAINING PROTEIN FROM BACILLUS SUBTILIS, NMR, 25 STRUCTURES | | Descriptor: | HISTIDINE CONTAINING PROTEIN | | Authors: | Jones, B.E, Rajagopal, P, Klevit, R.E. | | Deposit date: | 1997-04-01 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation on histidine is accompanied by localized structural changes in the phosphocarrier protein, HPr from Bacillus subtilis.
Protein Sci., 6, 1997
|
|
3J07
 
 | | Model of a 24mer alphaB-crystallin multimer | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Vollmar, B, Bardiaux, B, Dove, K.K, Rajagopal, P, Gonen, T, Oschkinat, H, Klevit, R.E. | | Deposit date: | 2011-04-27 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (20 Å), SOLID-STATE NMR, SOLUTION SCATTERING | | Cite: | N-terminal domain of {alpha}B-crystallin provides a conformational switch for multimerization and structural heterogeneity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2HID
 
 | |
1BSJ
 
 | | COBALT DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
2K31
 
 | | Solution Structure of cGMP-binding GAF domain of Phosphodiesterase 5 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, Phosphodiesterase 5A, cGMP-specific | | Authors: | Heikaus, C.C, Stout, J.R, Sekharan, M.R, Eakin, C.M, Rajagopal, P, Brzovic, P.S, Beavo, J.A, Klevit, R.E. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the cGMP Binding GAF Domain from Phosphodiesterase 5: Insights into Nucleotide Selectivity, Dimerization, and cGMP-Dependent Conformational Change.
J.Biol.Chem., 283, 2008
|
|
2JRF
 
 | | Solution NMR structure of Tubulin polymerization-promoting protein family member 3 from Homo sapiens. Northeast Structural Genomics target HR387. | | Descriptor: | Tubulin polymerization-promoting protein family member 3 | | Authors: | Aramini, J.M, Rossi, P, Shastry, R, Nwosu, C, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Rajan, P.K, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Tubulin polymerization-promoting protein family member 3 from Homo sapiens.
To be Published
|
|
2LDL
 
 | | Solution NMR Structure of the HIV-1 Exon Splicing Silencer 3 | | Descriptor: | RNA (27-MER) | | Authors: | Mishler, C, Levengood, J.D, Johnson, C.A, Rajan, P, Znosko, B.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Exon Splicing Silencer 3.
J.Mol.Biol., 415, 2012
|
|
9EY3
 
 | | The FK1 domain of FKBP51 in complex with (3S,11S,11aS)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxylic acid | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxylic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding proteins.
Chemistry, 2024
|
|
9EY4
 
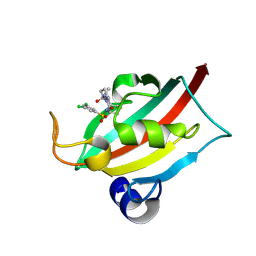 | | The FK1 domain of FKBP51 in complex with (3S,11S)-12-((3,5-dichlorophenyl)sulfonyl)-5-oxo-11-vinyldecahydro-1H-6,10-epiminopyrrolo[1,2-a]azonine-3-carboxamide | | Descriptor: | (1~{S},4~{S},7~{S},8~{S},9~{R})-13-[3,5-bis(chloranyl)phenyl]sulfonyl-8-ethenyl-2-oxidanylidene-3,13-diazatricyclo[7.3.1.0^{3,7}]tridecane-4-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Krajczy, P, Hausch, F. | | Deposit date: | 2024-04-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure-Based Design of Ultrapotent Tricyclic Ligands for FK506-Binding proteins.
Chemistry, 2024
|
|
1JM7
 
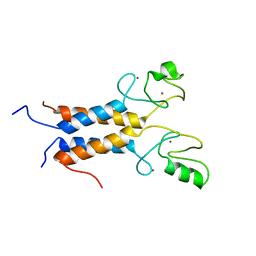 | | Solution structure of the BRCA1/BARD1 RING-domain heterodimer | | Descriptor: | BRCA1-ASSOCIATED RING DOMAIN PROTEIN 1, BREAST CANCER TYPE 1 SUSCEPTIBILITY PROTEIN, ZINC ION | | Authors: | Brzovic, P.S, Rajagopal, P, Hoyt, D.W, King, M.-C, Klevit, R.E. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.
Nat.Struct.Biol., 8, 2001
|
|
2KLR
 
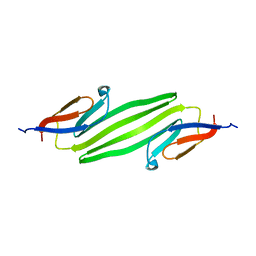 | | Solid-state NMR structure of the alpha-crystallin domain in alphaB-crystallin oligomers | | Descriptor: | Alpha-crystallin B chain | | Authors: | Jehle, S, Rajagopal, P, Markovic, S, Bardiaux, B, Kuehne, R, Higman, V.A, Klevit, R.E, van Rossum, B, Oschkinat, H. | | Deposit date: | 2009-07-08 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1DFF
 
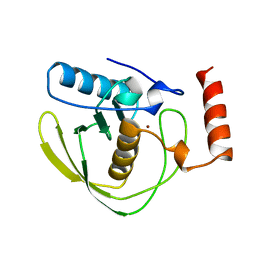 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
1BG5
 
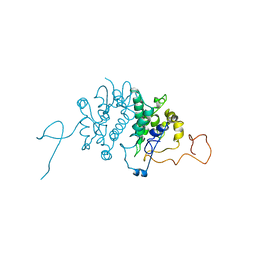 | | CRYSTAL STRUCTURE OF THE ANKYRIN BINDING DOMAIN OF ALPHA-NA,K-ATPASE AS A FUSION PROTEIN WITH GLUTATHIONE S-TRANSFERASE | | Descriptor: | FUSION PROTEIN OF ALPHA-NA,K-ATPASE WITH GLUTATHIONE S-TRANSFERASE | | Authors: | Zhang, Z, Devarajan, P, Morrow, J.S. | | Deposit date: | 1998-06-05 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the ankyrin-binding domain of alpha-Na,K-ATPase.
J.Biol.Chem., 273, 1998
|
|
1LO6
 
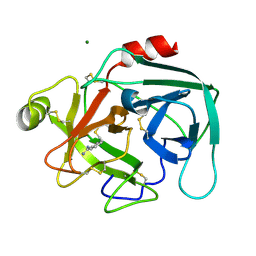 | | Human Kallikrein 6 (hK6) active form with benzamidine inhibitor at 1.56 A resolution | | Descriptor: | BENZAMIDINE, Kallikrein 6, MAGNESIUM ION | | Authors: | Bernett, M.J, Blaber, S.I, Scarisbrick, I.A, Dhanarajan, P, Thompson, S.M, Blaber, M. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure and biochemical characterization of human kallikrein 6 reveals a
trypsin-like kallikrein is expressed in the central nervous system
To be Published
|
|
1KKG
 
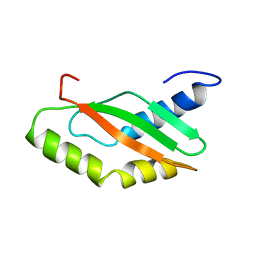 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
