2ZKH
 
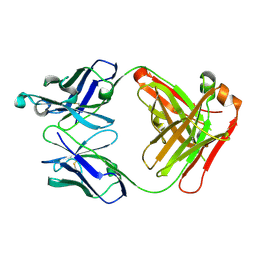 | | Human thrombopoietin neutralizing antibody TN1 FAB | | Descriptor: | Monoclonal TN1 FAB heavy chain, Monoclonal TN1 FAB light chain | | Authors: | Arai, S, Tamada, T, Honjo, E, Maeda, Y, Kuroki, R. | | Deposit date: | 2008-03-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | An insight into the thermodynamic characteristics of human thrombopoietin complexation with TN1 antibody.
Protein Sci., 25, 2016
|
|
5Y7Y
 
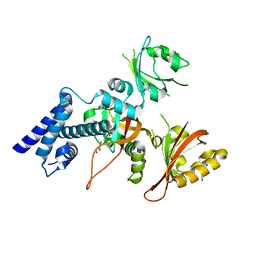 | | Crystal structure of AhRR/ARNT complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | Authors: | Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
4L1D
 
 | | Voltage-gated sodium channel beta3 subunit Ig domain | | Descriptor: | Sodium channel subunit beta-3 | | Authors: | Namadurai, S, Weimhofer, M, Rajappa, R, Stott, K, Klingauf, J, Chirgadze, D.Y, Jackson, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Molecular Imaging of the Nav Channel beta 3 Subunit Indicates a Trimeric Assembly.
J.Biol.Chem., 289, 2014
|
|
3WBH
 
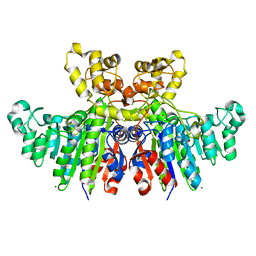 | | Structural characteristics of alkaline phosphatase from a moderately halophilic bacteria Halomonas sp.593 | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Ishibashi, M, Matsumoto, F, Tamada, T, Tokunaga, H, Tokunaga, M, Kuroki, R. | | Deposit date: | 2013-05-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characteristics of alkaline phosphatase from the moderately halophilic bacterium Halomonas sp. 593.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WRZ
 
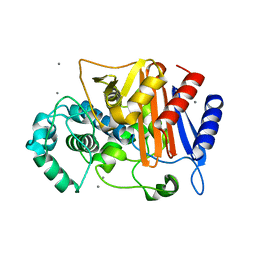 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (without soaking) | | Descriptor: | Beta-lactamase, CALCIUM ION, CHLORIDE ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WS0
 
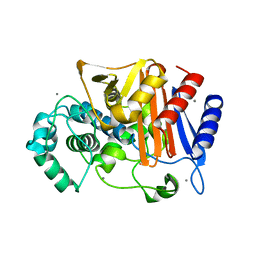 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-1A) | | Descriptor: | Beta-lactamase, CALCIUM ION, CESIUM ION, ... | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6KIL
 
 | | N21Q mutant thioredoxin from Halobacterium salinarum NRC-1 | | Descriptor: | Thioredoxin | | Authors: | Arai, S, Shibazaki, C, Shimizu, R, Adachi, M, Ishibashi, M, Tokunaga, H, Tokunaga, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic mechanism and evolutional characteristics of thioredoxin from Halobacterium salinarum NRC-1.
Acta Crystallogr.,Sect.D, 76, 2020
|
|
3VR2
 
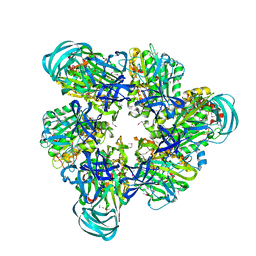 | | Crystal structure of nucleotide-free A3B3 complex from Enterococcus hirae V-ATPase [eA3B3] | | Descriptor: | V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR6
 
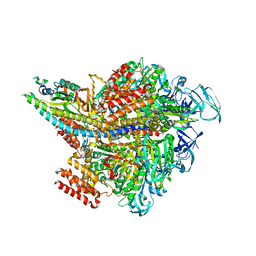 | | Crystal structure of AMP-PNP bound Enterococcus hirae V1-ATPase [bV1] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
3VR3
 
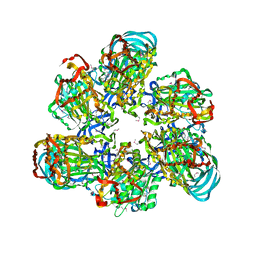 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
6TPF
 
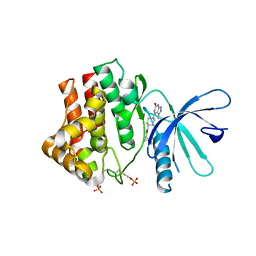 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | (1~{S})-2,2-bis(fluoranyl)-~{N}-[4-(3-methyl-6-oxidanylidene-2,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]cyclopropane-1-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Griessner, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6TPD
 
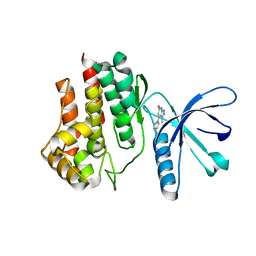 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 3-methyl-4-phenyl-2,7-dihydropyrazolo[3,4-b]pyridin-6-one, Tyrosine-protein kinase JAK2 | | Authors: | Hansen, B.B, Jepsen, T.J, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Moelck, C, Rai, S, Nasipireddy, V.R, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6TPE
 
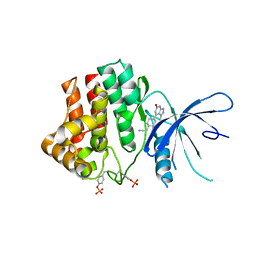 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 2-[4-(3-methyl-6-oxidanylidene-1,7-dihydropyrazolo[3,4-b]pyridin-4-yl)cyclohexyl]ethanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Hansen, B.B, Jepsen, T.H, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Molck, C, Rai, S, Nasipireddy, V.R, Jestel, A, Lammens, A, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
8YBK
 
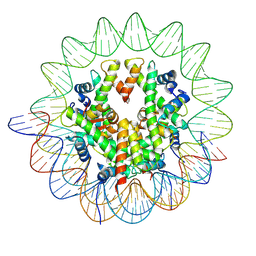 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
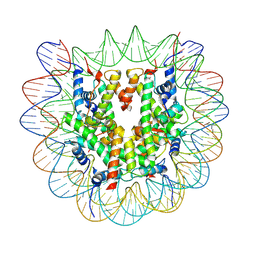 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
5J36
 
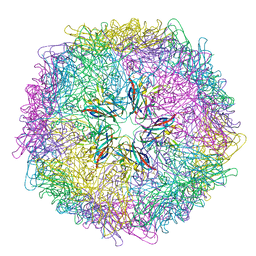 | | Crystal structure of 60-mer BFDV Capsid Protein | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
5J09
 
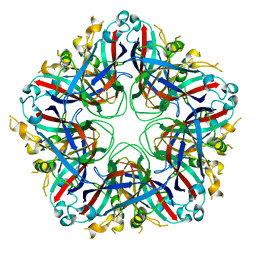 | |
7C0M
 
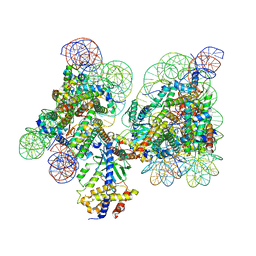 | | Human cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Kujirai, T, Zierhut, C, Takizawa, Y, Kim, R, Negishi, L, Uruma, N, Hirai, S, Funabiki, H, Kurumizaka, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the inhibition of cGAS by nucleosomes.
Science, 370, 2020
|
|
1WOE
 
 | | X-ray structure of a Z-DNA hexamer d(CGCGCG) | | Descriptor: | SPERMINE, Z-DNA hexamer | | Authors: | Chatake, T, Tanaka, I, Umino, H, Arai, S, Niimura, N. | | Deposit date: | 2004-08-15 | | Release date: | 2005-07-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7WLR
 
 | | Cryo-EM structure of the nucleosome containing Komagataella pastoris histones | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Fukushima, Y, Hatazawa, S, Hirai, S, Kujirai, T, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-01-13 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural and biochemical analyses of the nucleosome containing Komagataella pastoris histones.
J.Biochem., 172, 2022
|
|
1HE7
 
 | | Human Nerve growth factor receptor TrkA | | Descriptor: | GLYCEROL, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Banfield, M, Robertson, A, Allen, S, Dando, J, Tyler, S, Bennett, G, Brain, S, Mason, G, Holden, P, Clarke, A, Naylor, R, Wilcock, G, Brady, R, Dawbarn, D. | | Deposit date: | 2000-11-20 | | Release date: | 2001-04-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and Structure of the Nerve Growth Factor Binding Site on Trka.
Biochem.Biophys.Res.Commun., 282, 2001
|
|
3VI2
 
 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
1WQ2
 
 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2HBH
 
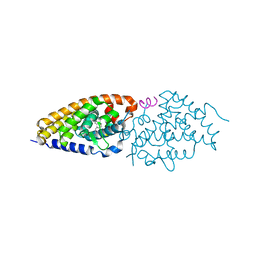 | | Crystal structure of Vitamin D nuclear receptor ligand binding domain bound to a locked side-chain analog of calcitriol and SRC-1 peptide | | Descriptor: | 1,3-CYCLOHEXANEDIOL, 4-METHYLENE-5-[(2E)-[(1S,3AS,7AS)-OCTAHYDRO-1-(5-HYDROXY-5-METHYL-1,3-HEXADIYNYL)-7A-METHYL-4H-INDEN-4-YLIDENE]ETHYLIDENE]-, (1R,3S,5Z), ... | | Authors: | Rochel, N, Hourai, S, Moras, D. | | Deposit date: | 2006-06-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the vitamin D nuclear receptor ligand binding domain in complex with a locked side chain analog of calcitriol
Arch.Biochem.Biophys., 460, 2007
|
|
