3T57
 
 | |
2AQ9
 
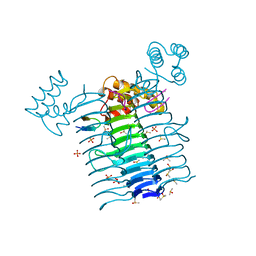 | | Structure of E. coli LpxA with a bound peptide that is competitive with acyl-ACP | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Williams, A.H, Immormino, R.M, Gewirth, D.T, Raetz, C.R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine acyltransferase with a bound antibacterial pentadecapeptide.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3HSQ
 
 | |
5YKA
 
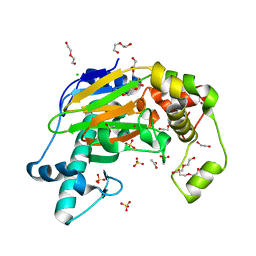 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with cobalt(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-10-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5YVZ
 
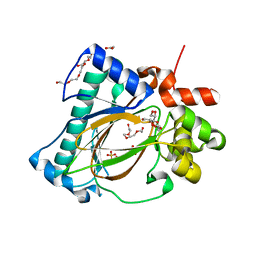 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and Fe(III) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5YW0
 
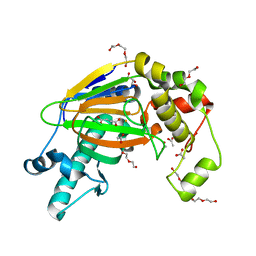 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with succinate and Fe(III) | | Descriptor: | ACETATE ION, FE (III) ION, GLYCEROL, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-11-28 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and Structural Insights into an Fe(II)/ alpha-Ketoglutarate/O2-Dependent Dioxygenase, Kdo 3-Hydroxylase (KdoO).
J. Mol. Biol., 430, 2018
|
|
5Y9X
 
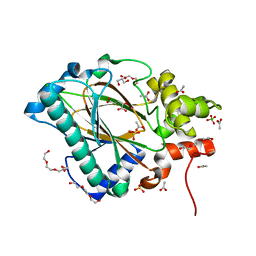 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and coblat(II) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-08-29 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
5Y9Y
 
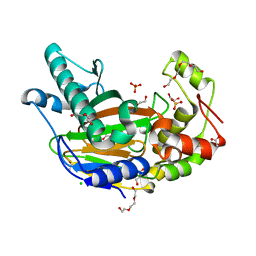 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with succinate and Co(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-08-29 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
4EHY
 
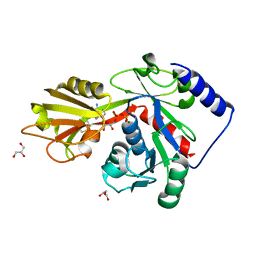 | | Crystal structure of LpxK from Aquifex aeolicus in complex with ADP/Mg2+ at 2.2 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JT2
 
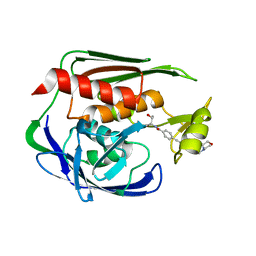 | | Solution Structure of the Aquifex aeolicus LpxC- CHIR-090 complex | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Barb, A.W, Jiang, L, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2007-07-18 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the deacetylase LpxC bound to the antibiotic CHIR-090: Time-dependent inhibition and specificity in ligand binding
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4EHW
 
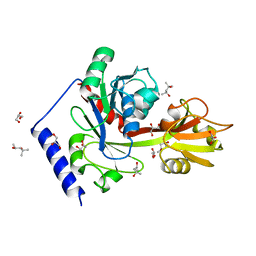 | | Crystal structure of LpxK from Aquifex aeolicus at 2.3 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Tetraacyldisaccharide 4'-kinase | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EHX
 
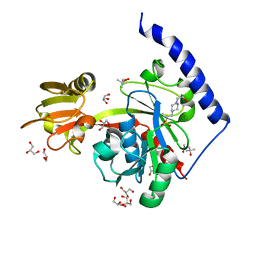 | | Crystal structure of LpxK from Aquifex aeolicus at 1.9 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Emptage, R.P, Daughtry, K.D, Pemble IV, C.W, Raetz, C.R.H. | | Deposit date: | 2012-04-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of LpxK, the 4'-kinase of lipid A biosynthesis and atypical P-loop kinase functioning at the membrane interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3EH0
 
 | | Crystal Structure of LpxD from Escherichia coli | | Descriptor: | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Bartling, C.M, Raetz, C.R.H. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis
Biochemistry, 48, 2009
|
|
4ITM
 
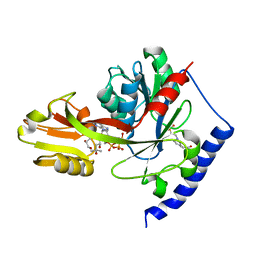 | | Crystal structure of "apo" form LpxK from Aquifex aeolicus in complex with ATP at 2.2 angstrom resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Emptage, R.P, Pemble IV, C.W, York, J.D, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1994 Å) | | Cite: | Mechanistic Characterization of the Tetraacyldisaccharide-1-phosphate 4'-Kinase LpxK Involved in Lipid A Biosynthesis.
Biochemistry, 52, 2013
|
|
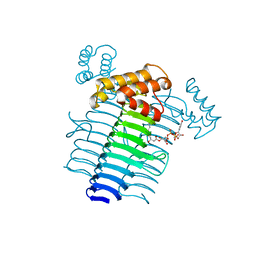
2QIV
 
 | |
2QIA
 
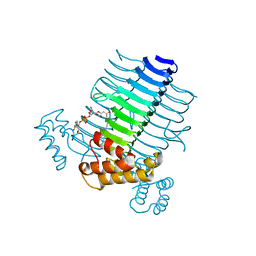 | |
1XXE
 
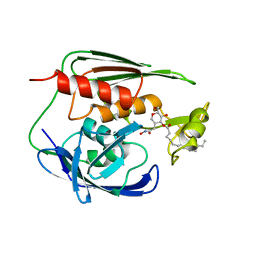 | | RDC refined solution structure of the AaLpxC/TU-514 complex | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Coggins, B.E, McClerren, A.L, Jiang, L, Li, X, Rudolph, J, Hindsgaul, O, Raetz, C.R.H, Zhou, P. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the LpxC-TU-514 Complex and pK(a) Analysis of an Active Site Histidine: Insights into the Mechanism and Inhibitor Design
Biochemistry, 44, 2005
|
|
1YUC
 
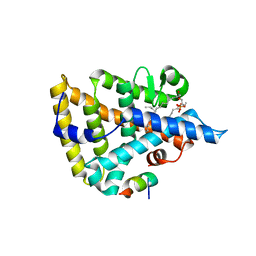 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to Phospholipid and a Fragment of Human SHP | | Descriptor: | GLYCEROL, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor 0B2, ... | | Authors: | Ortlund, E.A, Yoonkwang, L, Solomon, I.H, Hager, J.M, Safi, R, Choi, Y, Guan, Z, Tripathy, A, Raetz, C.R.H, McDonnell, D.P, Moore, D.D, Redinbo, M.R. | | Deposit date: | 2005-02-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP
Nat.Struct.Mol.Biol., 12, 2005
|
|
1MM4
 
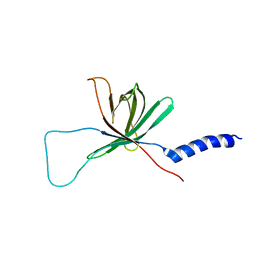 | | Solution NMR structure of the outer membrane enzyme PagP in DPC micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MM5
 
 | | Solution NMR structure of the outer membrane enzyme PagP in OG micelles | | Descriptor: | CrcA protein | | Authors: | Hwang, P.M, Choy, W.-Y, Lo, E.I, Chen, L, Forman-Kay, J.D, Raetz, C.R.H, Prive, G.G, Bishop, R.E, Kay, L.E. | | Deposit date: | 2002-09-03 | | Release date: | 2002-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Outer Membrane Enzyme PagP by NMR
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2BLL
 
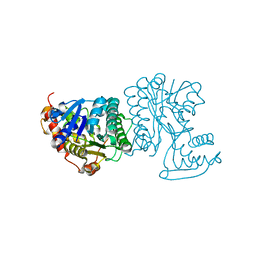 | | Apo-structure of the C-terminal decarboxylase domain of ArnA | | Descriptor: | PROTEIN YFBG | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L- Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
2BLN
 
 | | N-terminal formyltransferase domain of ArnA in complex with N-5- formyltetrahydrofolate and UMP | | Descriptor: | ACETATE ION, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PROTEIN YFBG, ... | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L-Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
4IHH
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHF
 
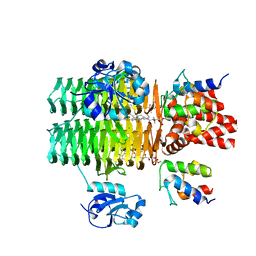 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | Acyl carrier protein, CHLORIDE ION, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxytetradecanethioate, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
4IHG
 
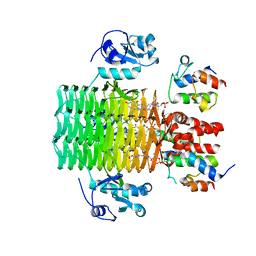 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
