1CDU
 
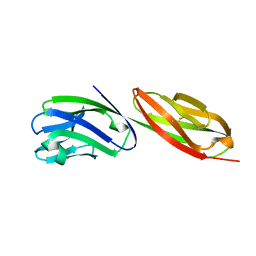 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH PHE 43 REPLACED BY VAL | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1C81
 
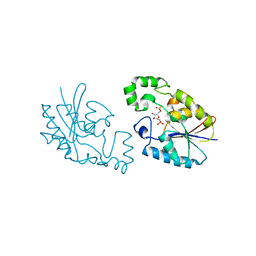 | | MICHAELIS COMPLEX OF FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1-deoxy-1-phosphono-6-O-phosphono-D-glucitol, FRUCTOSE-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.H, Olson, T.W, McClard, R.W, Witte, J.F, McFarlan, S.C, Banaszak, L.J, Levitt, D.G, Lange, A.J. | | Deposit date: | 2000-04-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction Mechanism of Fructose-2,6-bisphosphatase Suggested by the Crystal Structures of a pseudo-Michaelis complex and Metabolite Complexes
To be Published
|
|
1CDI
 
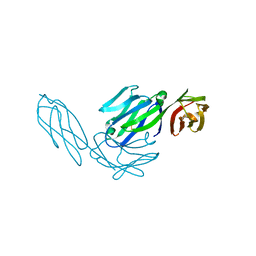 | |
1CDY
 
 | | STRUCTURE OF T-CELL SURFACE GLYCOPROTEIN CD4 MUTANT WITH GLY 47 REPLACED BY SER | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD4 | | Authors: | Wu, H, Myszka, D, Tendian, S.W, Brouillette, C.G, Sweet, R.W, Chaiken, I.M, Hendrickson, W.A. | | Deposit date: | 1996-11-11 | | Release date: | 1997-04-01 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural analysis of mutant CD4 receptors that are defective in HIV gp120 binding.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3F1L
 
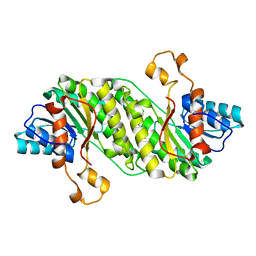 | |
3F4W
 
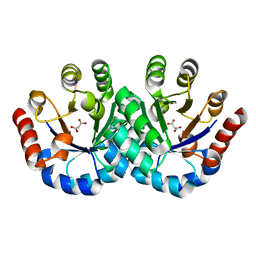 | |
5JVE
 
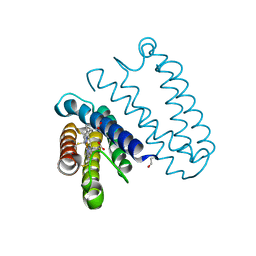 | |
5JW3
 
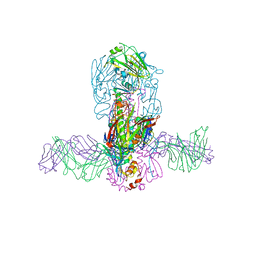 | | Structure of MEDI8852 Fab Fragment in Complex with H7 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Collins, P.J, Neu, U, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
8C54
 
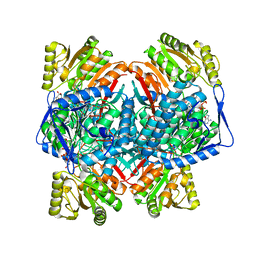 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5JSL
 
 | | The L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous form | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
8C12
 
 | | Identification of an intermediate activation state of PAK5 reveals a novel mechanism of kinase inhibition. | | Descriptor: | PAK5-Af17, Serine/threonine-protein kinase PAK 5 | | Authors: | Martin, H.L, Turner, A.L, Trinh, C.H, Bayliss, R.W, Tomlinson, D.C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Affimer-mediated locking of p21-activated kinase 5 in an intermediate activation state results in kinase inhibition.
Cell Rep, 42, 2023
|
|
5JW4
 
 | | Structure of MEDI8852 Fab Fragment in Complex with H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
4HTN
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 1.32 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5JT4
 
 | | L16A mutant of cytochrome c prime from Alcaligenes xylosoxidans: Ferrous state | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-09 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
5JS5
 
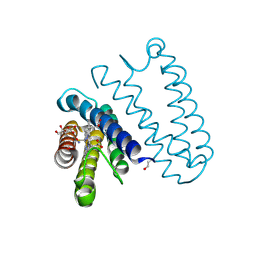 | | Nitric oxide complex of the L16F mutant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | ASCORBIC ACID, Cytochrome c', HEME C, ... | | Authors: | Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering proximal vs. distal heme-NO coordination via dinitrosyl dynamics: implications for NO sensor design.
Chem Sci, 8, 2017
|
|
5JUA
 
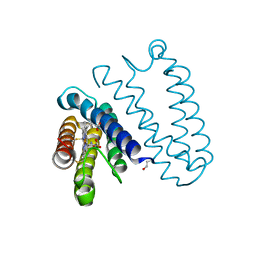 | |
3F1K
 
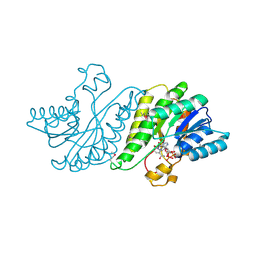 | |
4HTK
 
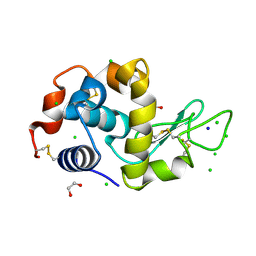 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 2.17 x 10e+12 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.A, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7LVZ
 
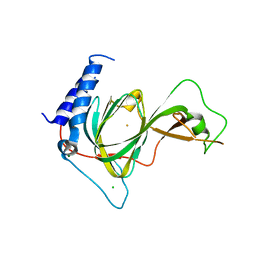 | | Crystal structure of ADO | | Descriptor: | 2-aminoethanethiol dioxygenase, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bingman, C.A, Fernandez, R.L, Smith, R.W, Fox, B.G, Brunold, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Crystal Structure of Cysteamine Dioxygenase Reveals the Origin of the Large Substrate Scope of This Vital Mammalian Enzyme.
Biochemistry, 60, 2021
|
|
8CUK
 
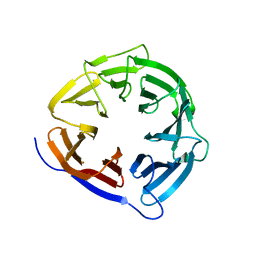 | |
3G4X
 
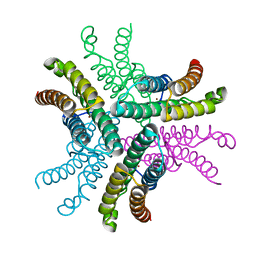 | | Crystal Structure of NiSOD Y9F mutant | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
8DAL
 
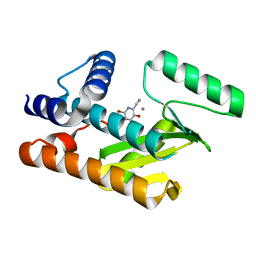 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinonitrile | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carbonitrile, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDB
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinic acid | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDE
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-2-(1H-tetrazol-5-yl) yridine-3-ol | | Descriptor: | (2M)-6-bromo-3-hydroxy-2-(1H-tetrazol-5-yl)pyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
