2B1Z
 
 | | Human estrogen receptor alpha ligand-binding domain in complex with 17methyl-17alpha-dihydroequilenin and a glucoc interacting protein 1 NR box II peptide | | Descriptor: | 17-METHYL-17-ALPHA-DIHYDROEQUILENIN, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Molecular characterization of a B-ring unsaturated estrogen: implications for conjugated equine estrogen components of premarin.
Steroids, 73, 2008
|
|
2K41
 
 | | NMR structure of uridine bulged RNA duplex | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*UP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2AOE
 
 | | crystal structure analysis of HIV-1 protease mutant V82A with a substrate analog CA-P2 | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2K3Z
 
 | | NMR structure of adenosine bulged RNA duplex with C:G-A triple | | Descriptor: | RNA_(5'-R(*CP*AP*GP*CP*CP*GP*AP*C)-3')_, RNA_(5'-R(*GP*UP*CP*GP*AP*GP*CP*UP*G)-3')_ | | Authors: | Popenda, L, Bielecki, L, Gdaniec, Z, Adamiak, R.W. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple
ARKIVOC, 2009, 2008
|
|
2JXQ
 
 | | NMR structure of RNA duplex | | Descriptor: | RNA (5'-R(*CP*GP*CP*UP*CP*UP*CP*UP*GP*C)-3'), RNA (5'-R(*GP*CP*AP*GP*AP*GP*AP*GP*CP*G)-3') | | Authors: | Popenda, L, Adamiak, R.W, Gdaniec, Z. | | Deposit date: | 2007-11-28 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Bulged adenosine influence on the RNA duplex conformation in solution.
Biochemistry, 47, 2008
|
|
2BDO
 
 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
2ANT
 
 | | THE 2.6 A STRUCTURE OF ANTITHROMBIN INDICATES A CONFORMATIONAL CHANGE AT THE HEPARIN BINDING SITE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose, ANTITHROMBIN | | Authors: | Skinner, R, Abrahams, J.-P, Whisstock, J.C, Lesk, A.M, Carrell, R.W, Wardell, M.R. | | Deposit date: | 1997-01-28 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 A structure of antithrombin indicates a conformational change at the heparin binding site.
J.Mol.Biol., 266, 1997
|
|
2AVV
 
 | | Kinetics, stability, and structural changes in high resolution crystal structures of HIV-1 protease with drug resistant mutations L24I, I50V, and G73S | | Descriptor: | ACETIC ACID, CHLORIDE ION, N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, ... | | Authors: | Liu, F, Boross, P.I, Wang, Y.F, Tozser, J, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S.
J.Mol.Biol., 354, 2005
|
|
2AOF
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P1-P6 | | Descriptor: | ACETIC ACID, CHLORIDE ION, PEPTIDE INHIBITOR, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2B6A
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with THR-50 | | Descriptor: | 1-(2,6-DIFLUOROBENZYL)-2-(2,6-DIFLUOROPHENYL)-4-METHYL-1H-BENZIMIDAZOLE, Reverse transcriptase p51 subunit, Reverse transcriptase p66 subunit | | Authors: | Morningstar, M.L, Roth, T, Smith, M.K, Zajac, M, Watson, K, Buckheit, R.W, Das, K, Zhang, W, Arnold, E, Michejda, C.J. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with THR-50
TO BE PUBLISHED
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | Descriptor: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2005-08-12 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
2BO0
 
 | | Crystal structure of the C130A mutant of nitrite reductase from Alcaligenes xylosoxidans | | Descriptor: | DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Hough, M.A, Ellis, M.J, Antonyuk, S, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-04-06 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structural Studies of Mutants Provide Insights Into Catalysis and Electron Transfer Processes in Copper Nitrite Reductase
J.Mol.Biol., 350, 2005
|
|
2N0W
 
 | | Mdmx-SJ212 | | Descriptor: | 4-({(4S,5R)-4-(5-bromo-2-fluorophenyl)-5-(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N06
 
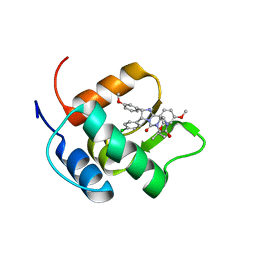 | | Mdmx-298 | | Descriptor: | 4-[[(4S,5R)-5-(4-chlorophenyl)-4-(3-methoxyphenyl)-2-(4-methoxy-2-propan-2-yloxy-phenyl)-4,5-dihydroimidazol-1-yl]carbonyl]piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
1YC6
 
 | |
2JLP
 
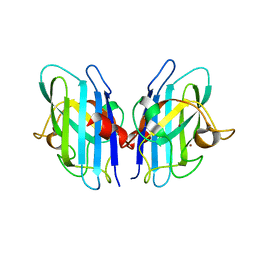 | | Crystal structure of human extracellular copper-zinc superoxide dismutase. | | Descriptor: | COPPER (II) ION, EXTRACELLULAR SUPEROXIDE DISMUTASE (CU-ZN), THIOCYANATE ION, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Marklund, S.L, Hasnain, S.S. | | Deposit date: | 2008-09-14 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Human Extracellular Copper-Zinc Superoxide Dismutase at 1.7 A Resolution: Insights Into Heparin and Collagen Binding.
J.Mol.Biol., 388, 2009
|
|
2JUG
 
 | |
2K47
 
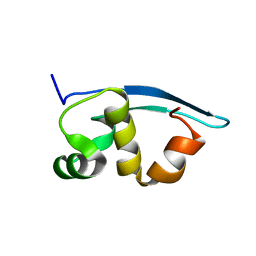 | | Solution structure of the C-terminal N-RNA binding domain of the Vesicular Stomatitis Virus Phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Ribeiro, E.A, Favier, A, Gerard, F.C, Leyrat, C, Brutscher, B, Blondel, D, Ruigrok, R.W, Blackledge, M, Jamin, M. | | Deposit date: | 2008-05-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the C-Terminal Nucleoprotein-RNA Binding Domain of the Vesicular Stomatitis Virus Phosphoprotein.
J.Mol.Biol., 2008
|
|
1XZ4
 
 | | Intersubunit Interactions Associated with Tyr42alpha Stabilize the Quaternary-T Tetramer but are not Major Quaternary Constraints in Deoxyhemoglobin: alphaY42A deoxyhemoglobin no-salt | | Descriptor: | Hemoglobin alpha chain, Hemoglobin beta chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Rogers, P.H, Arnone, A, Hui, H.L, Wierzba, A, DeYoung, A, Kwiatkowski, L.D, Noble, R.W, Juszczak, L.J, Peterson, E.S, Friedman, J.M. | | Deposit date: | 2004-11-11 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intersubunit interactions associated with tyr42alpha stabilize the quaternary-T tetramer but are not major quaternary constraints in deoxyhemoglobin
Biochemistry, 44, 2005
|
|
1XYS
 
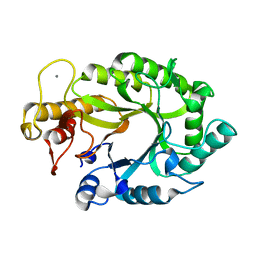 | | CATALYTIC CORE OF XYLANASE A E246C MUTANT | | Descriptor: | CALCIUM ION, XYLANASE A | | Authors: | Harris, G.W, Jenkins, J.A, Connerton, I, Pickersgill, R.W. | | Deposit date: | 1994-09-02 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites.
Structure, 2, 1994
|
|
2LBZ
 
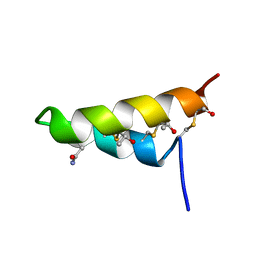 | | Thurincin H | | Descriptor: | Thuricin17 | | Authors: | Sit, C.S, van Belkum, M.J, Mckay, R.T, Worobo, R.W, Vederas, J.C. | | Deposit date: | 2011-04-10 | | Release date: | 2012-01-18 | | Last modified: | 2018-08-22 | | Method: | SOLUTION NMR | | Cite: | The 3D solution structure of thurincin H, a bacteriocin with four sulfur to alpha-carbon crosslinks.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2C53
 
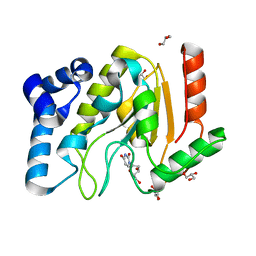 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | Authors: | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2C56
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | URACIL DNA GLYCOSYLASE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Krusong, K, Carpenter, E.P, Bellamy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2N0U
 
 | | Mdmx-057 | | Descriptor: | 4-[(4S,5R)-4-(3-chlorophenyl)-5-(4-chlorophenyl)-1-(3-oxidanylidenepiperazin-1-yl)carbonyl-4,5-dihydroimidazol-2-yl]-3-propan-2-yloxy-benzenecarbonitrile, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2BWI
 
 | | Atomic Resolution Structure of Nitrite -soaked Achromobacter cycloclastes Cu Nitrite Reductase | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Strange, R.W, Sawers, G, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2005-07-14 | | Release date: | 2005-08-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structures of Resting-State, Substrate- and Product-Complexed Cu-Nitrite Reductase Provide Insight Into Catalytic Mechanism
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
