4W4S
 
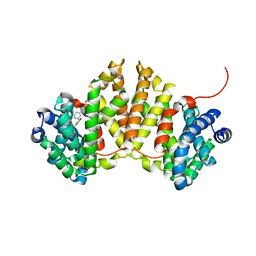 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum in complex with BPH-629 | | Descriptor: | Uncharacterized protein blr2150, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
830C
 
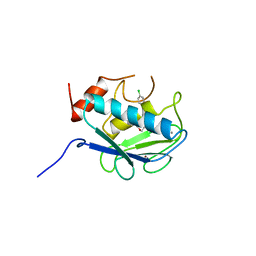 | | COLLAGENASE-3 (MMP-13) COMPLEXED TO A SULPHONE-BASED HYDROXAMIC ACID | | Descriptor: | 4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYLMETHYL]-TETRAHYDRO-PYRAN-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, MMP-13, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-06 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
7JR6
 
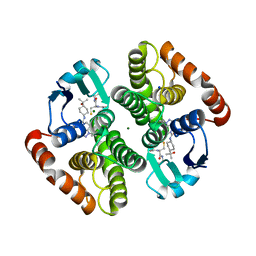 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR8
 
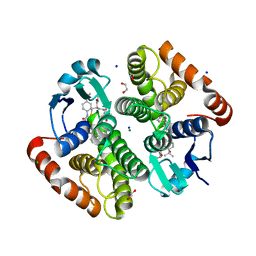 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
4I4L
 
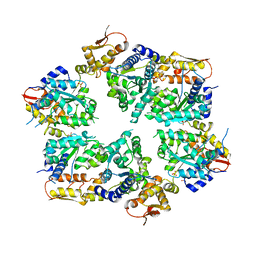 | | Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | Authors: | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6981 Å) | | Cite: | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
6CHR
 
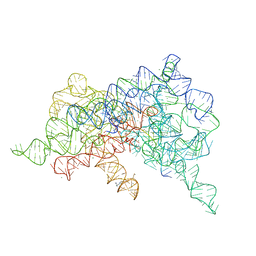 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
5V1Z
 
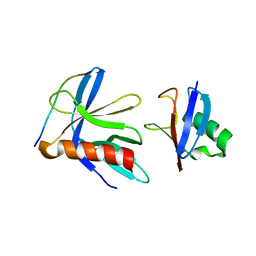 | | Crystal structure of the RPN13 PRU-RPN2 (932-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
8IH4
 
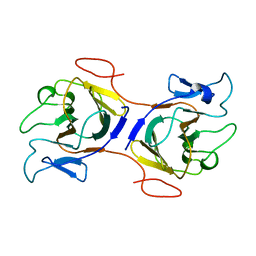 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A2 Mutant | | Descriptor: | Butyrophilin subfamily 2 member A2 | | Authors: | Yang, Y.Y, Yi, S.M, Zhang, M.T, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-22 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IGT
 
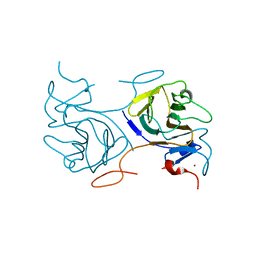 | | Crystal Structure of Intracellular B30.2 Domain of BTN2A1 | | Descriptor: | Butyrophilin subfamily 2 member A1, ZINC ION | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2023-02-21 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
6YSE
 
 | |
5KPY
 
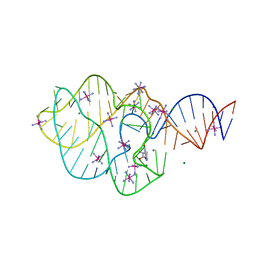 | | Structure of a 5-hydroxytryptophan aptamer | | Descriptor: | 5-hydroxy-L-tryptophan, 5-hydroxytryptophan RNA aptamer, IRIDIUM HEXAMMINE ION, ... | | Authors: | Batey, R.T, Porter, E, Merck, M. | | Deposit date: | 2016-07-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors.
Nat. Chem. Biol., 13, 2017
|
|
3EOD
 
 | |
3ES2
 
 | |
7QPB
 
 | | Catalytic C-lobe of the HECT-type ubiquitin ligase E6AP in complex with a hybrid foldamer-peptide macrocycle | | Descriptor: | Isoform I of Ubiquitin-protein ligase E3A, hybrid foldamer-peptide macrocycle | | Authors: | Dengler, S, Howard, R.T, Morozov, V, Tsiamantas, C, Douat, C, Suga, H, Huc, I. | | Deposit date: | 2022-01-03 | | Release date: | 2023-09-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Display Selection of a Hybrid Foldamer-Peptide Macrocycle.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7SJY
 
 | |
5CUW
 
 | |
5D4Y
 
 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, xylanase | | Authors: | Zheng, Y, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5CUY
 
 | | Crystal structure of Trypanosoma brucei Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, CITRIC ACID, MAGNESIUM ION | | Authors: | Yang, Y.Y, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
5DBF
 
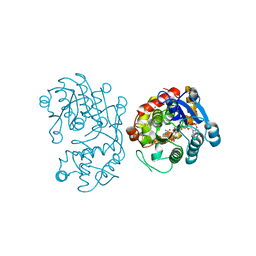 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NADPH | | Descriptor: | Iridoid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CUU
 
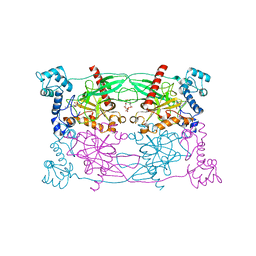 | | Crystal structure of Trypanosoma cruzi Vacuolar Soluble Pyrophosphatases in complex with bisphosphonate inhibitor BPH-1260 | | Descriptor: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Acidocalcisomal pyrophosphatase, D-MALATE, ... | | Authors: | Liu, W.D, Yang, Y.Y, Ko, T.P, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
5DBI
 
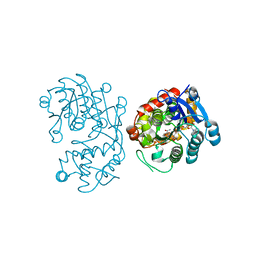 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ and 10-oxogeranial | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6ITA
 
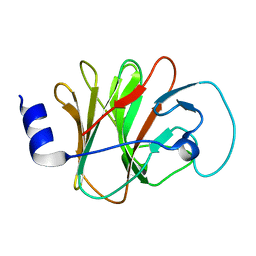 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6ISM
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J0L
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with sulfate ion | | Descriptor: | Butyrophilin subfamily 3 member A3, SULFATE ION | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
