1DWS
 
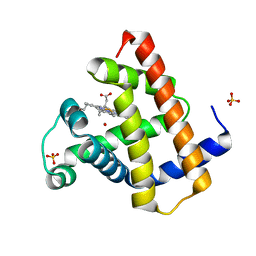 | | PHOTOLYZED CARBONMONOXY MYOGLOBIN (HORSE HEART) | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1T60
 
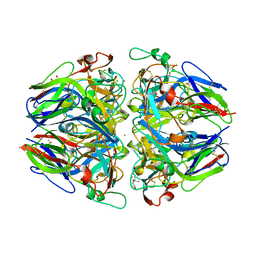 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
1DWR
 
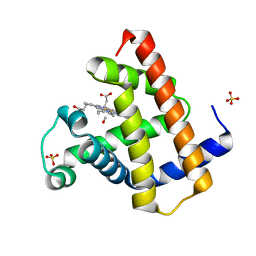 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH CO | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-11 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1ST9
 
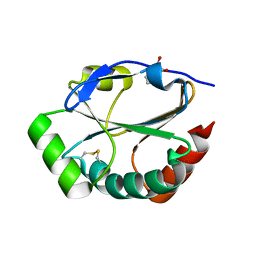 | | Crystal Structure of a Soluble Domain of ResA in the Oxidised Form | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A, Acheson, R.M, Le Brun, N.E, Oubrie, A. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Redox-coupled Protein Substrate Selection by the Cytochrome c Biosynthesis Protein ResA.
J.Biol.Chem., 279, 2004
|
|
1CER
 
 | |
1E2B
 
 | | NMR STRUCTURE OF THE C10S MUTANT OF ENZYME IIB CELLOBIOSE OF THE PHOSPHOENOL-PYRUVATE DEPENDENT PHOSPHOTRANSFERASE SYSTEM OF ESCHERICHIA COLI, 17 STRUCTURES | | Descriptor: | ENZYME IIB-CELLOBIOSE | | Authors: | Ab, E, Schuurman-Wolters, G, Reizer, J, Saier, M.H, Dijkstra, K, Scheek, R.M, Robillard, G.T. | | Deposit date: | 1996-11-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR side-chain assignments and solution structure of enzyme IIBcellobiose of the phosphoenolpyruvate-dependent phosphotransferase system of Escherichia coli.
Protein Sci., 6, 1997
|
|
1DWT
 
 | | Photorelaxed horse heart MYOGLOBIN CO complex | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chu, K, Vojtechovsky, J, McMahon, B.H, Sweet, R.M, Berendzen, J, Schlichting, I. | | Deposit date: | 1999-12-12 | | Release date: | 2000-03-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a New Ligand Binding Intermediate in Wildtype Carbonmonoxy Myoglobin
Nature, 403, 2000
|
|
1W8X
 
 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1DXD
 
 | | Photolyzed CO complex of Myoglobin Mb-YQR at 20K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Vallone, B, Cutruzzola, F, Travaglini-Allocatelli, C, Berendzen, J, Chu, K, Sweet, R.M, Schlichting, I. | | Deposit date: | 2000-01-03 | | Release date: | 2000-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Role of Cavities in Protein Dynamics: Crystal Structure of a Photolytic Intermediate of a Mutant Myoglobin.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5WIU
 
 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5WPR
 
 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WAX
 
 | | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10) | | Descriptor: | GLYCEROL, SAPK10 (serine/threonine-protein kinase 10) | | Authors: | Righetto, G.L, Counago, R.M, Halabelian, L, Santiago, A.S, Massirer, K.B, Arruda, P, Gileadi, O, Menossi, M, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10)
To Be Published
|
|
1M4N
 
 | | CRYSTAL STRUCTURE OF APPLE ACC SYNTHASE IN COMPLEX WITH [2-(AMINO-OXY)ETHYL](5'-DEOXYADENOSIN-5'-YL)(METHYL)SULFONIUM | | Descriptor: | (2-AMINOOXY-ETHYL)-[5-(6-AMINO-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDRO-FURAN-2-YLMETHYL]-METHYL-SULFONIUM, 1-aminocyclopropane-1-carboxylate synthase, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Capitani, G, Eliot, A.C, Gut, H, Khomutov, R.M, Kirsch, J.F, Grutter, M.G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase in complex with an amino-oxy analogue of the substrate: implications for substrate binding.
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1647, 2003
|
|
5WNA
 
 | | Structure of antibody 3D3 bound to the central conserved region of RSV G | | Descriptor: | Major surface glycoprotein G, mAb 3D3 Fab heavy chain, mAb 3D3 Fab light chain | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
5WPP
 
 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
1I53
 
 | | RE(I)-TRICARBONYL DIIMINE (Q107H)) AZURIN | | Descriptor: | 4,7-DIMETHYL-[1,10]PHENANTHROLINE, AZURIN, COPPER (II) ION, ... | | Authors: | Di Bilio, A.J, Crane, B.R, Wehbi, W.A, Kiser, C.N, Abu-Omar, M.M, Carlos, R.M, Richards, J.H, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-02-24 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Properties of photogenerated tryptophan and tyrosyl radicals in structurally characterized proteins containing rhenium(I) tricarbonyl diimines.
J.Am.Chem.Soc., 123, 2001
|
|
1N23
 
 | | (+)-Bornyl diphosphate synthase: Complex with Mg, pyrophosphate, and (1R,4S)-2-azabornane | | Descriptor: | (+)-bornyl diphosphate synthase, (1R,4S)-2-AZABORNANE, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5W7R
 
 | | Crystal structure of OxaC in complex with SAH and oxaline | | Descriptor: | (3E,7aR,12aS)-3-[(1H-imidazol-4-yl)methylidene]-6,12-dimethoxy-7a-(2-methylbut-3-en-2-yl)-7a,12-dihydro-1H,5H-imidazo[1 ',2':1,2]pyrido[2,3-b]indole-2,5(3H)-dione, OxaC, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Newmister, S.A, Romminger, S, Schmidt, J.J, Williams, R.M, Smith, J.L, Berlinck, R.G.S, Sherman, D.H. | | Deposit date: | 2017-06-20 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Unveiling sequential late-stage methyltransferase reactions in the meleagrin/oxaline biosynthetic pathway.
Org. Biomol. Chem., 16, 2018
|
|
5WPU
 
 | | Crystal structure HpiC1 Y101S | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
1N24
 
 | | (+)-Bornyl diphosphate synthase: Complex with Mg and product | | Descriptor: | (+)-BORNYL DIPHOSPHATE, (+)-bornyl diphosphate synthase, MAGNESIUM ION | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IIR
 
 | | Crystal Structure of UDP-glucosyltransferase GtfB | | Descriptor: | MAGNESIUM ION, SULFATE ION, glycosyltransferase GtfB | | Authors: | Mulichak, A.M, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics.
Structure, 9, 2001
|
|
5TE0
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120 | | Descriptor: | AP2-associated protein kinase 1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Counago, R.M, Elkins, J.M, Bountra, C, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120
To Be Published
|
|
8P37
 
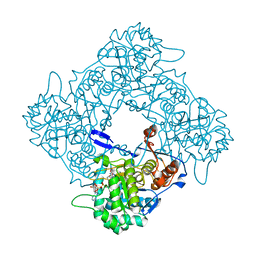 | | Structure a catalytically inactive mutant of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
8P4Q
 
 | | Structure of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, INOSINIC ACID, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
6P6E
 
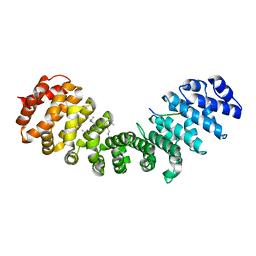 | | Structure of Mouse Importin alpha - PAC3 NLS peptide complex | | Descriptor: | Importin subunit alpha-1, PAC3 NLS | | Authors: | Bernardes, N.E, Silva, T.D, Fukuda, C.A, Oliveira, H.C, Fontes, M.R.M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Comparative study of the interactions between fungal transcription factor nuclear localization sequences with mammalian and fungal importin-alpha.
Sci Rep, 10, 2020
|
|
