7LSO
 
 | | L-Phenylseptin | | Descriptor: | L-Phenylseptin peptide | | Authors: | Munhoz, V.H, Verly, R.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of L-Phenylseptin in DPC-d38.
To Be Published
|
|
7LSP
 
 | |
3LPW
 
 | | Crystal structure of the FnIII-tandem A77-A78 from the A-band of titin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, A77-A78 domain from Titin | | Authors: | Bucher, R.M, Mayans, O. | | Deposit date: | 2010-02-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the FnIII Tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin
J.Mol.Biol., 401, 2010
|
|
7MTO
 
 | |
3LPA
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LBS
 
 | |
3LPD
 
 | | Crystal structure of a subtilisin-like protease | | Descriptor: | Acidic extracellular subtilisin-like protease AprV2, CALCIUM ION | | Authors: | Porter, C.J, Wong, W, Whisstock, J.C, Rood, J.I, Kennan, R.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Subtilisin-Like Protease AprV2 Is Required for Virulence and Uses a Novel Disulphide-Tethered Exosite to Bind Substrates
Plos Pathog., 6, 2010
|
|
3LZ9
 
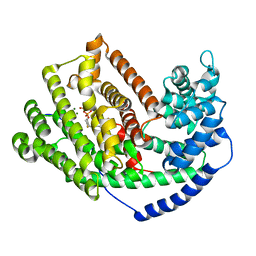 | | The Crystal Structure of 5-epi-aristolochene synthase M4 mutant complexed with (2-trans,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
7MKE
 
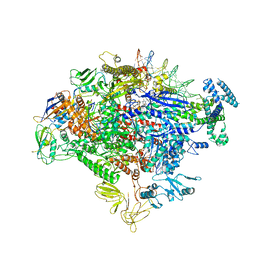 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MMM
 
 | | LyeTx I | | Descriptor: | Toxin LyeTx 1 | | Authors: | Verly, R.M. | | Deposit date: | 2021-04-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | LyeTx I, a potent antimicrobial peptide from the venom of the spider Lycosa erythrognatha
Amino Acids, 39, 2010
|
|
7MKI
 
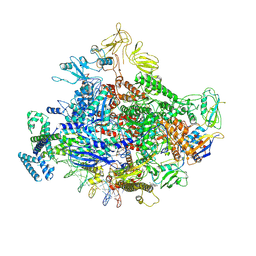 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3MLH
 
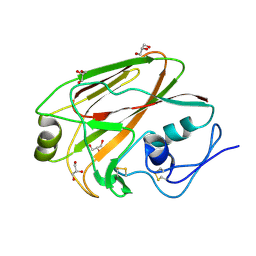 | | Crystal structure of the 2009 H1N1 influenza virus hemagglutinin receptor-binding domain | | Descriptor: | GLYCEROL, Hemagglutinin | | Authors: | DuBois, R.M, Aguilar-Yanez, J.M, Mendoza-Ochoa, G.I, Schultz-Cherry, S, Alvarez, M.M, White, S.W, Russell, C.J. | | Deposit date: | 2010-04-16 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The Receptor-Binding Domain of Influenza Virus Hemagglutinin Produced in Escherichia coli Folds into Its Native, Immunogenic Structure.
J.Virol., 85, 2011
|
|
3LZU
 
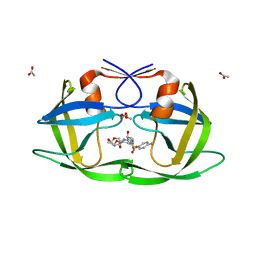 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
7MN3
 
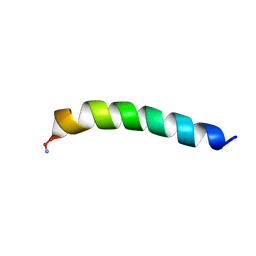 | | Homotarsinin monomer - Htr-M | | Descriptor: | Homotarsinin | | Authors: | Verly, R.M. | | Deposit date: | 2021-04-30 | | Release date: | 2021-07-14 | | Method: | SOLUTION NMR | | Cite: | Structure and membrane interactions of the homodimeric antibiotic peptide homotarsinin.
Sci Rep, 7, 2017
|
|
3LZC
 
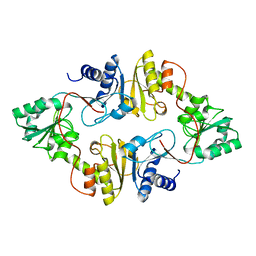 | | Crystal structure of Dph2 from Pyrococcus horikoshii | | Descriptor: | Dph2 | | Authors: | Zhang, Y, Zhu, X, Torelli, A.T, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3M00
 
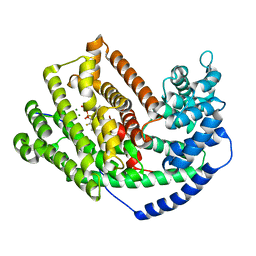 | | Crystal Structure of 5-epi-aristolochene synthase M4 mutant complexed with (2-cis,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
6CE2
 
 | | Crystal structure of Myotoxin I (MjTX-I) from Bothrops moojeni complexed to inhibitor suramin | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Basic phospholipase A2 homolog 1 | | Authors: | Salvador, G.H.M, Fontes, M.R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional characterization of suramin-bound MjTX-I from Bothrops moojeni suggests a particular myotoxic mechanism.
Sci Rep, 8, 2018
|
|
3LZD
 
 | | Crystal structure of Dph2 from Pyrococcus horikoshii with 4Fe-4S cluster | | Descriptor: | Dph2, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Torelli, A.T, Zhang, Y, Zhu, X, Lee, M, Dzikovski, B, Koralewski, R.M, Wang, E, Freed, J, Krebs, C, Lin, H, Ealick, S.E. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme.
Nature, 465, 2010
|
|
3M02
 
 | | The Crystal Structure of 5-epi-aristolochene synthase complexed with (2-cis,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, MAGNESIUM ION | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
3M21
 
 | | Crystal structure of DmpI from Helicobacter pylori Determined to 1.9 Angstroms resolution | | Descriptor: | Probable tautomerase HP_0924 | | Authors: | Hackert, M.L, Whitman, C.P, Almrud, J.J, Dasgupta, R, Kern, A.D, Czerwinski, R.M. | | Deposit date: | 2010-03-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural characterization of DmpI from Helicobacter pylori and Archaeoglobus fulgidus, two 4-oxalocrotonate tautomerase family members.
Bioorg.Chem., 38, 2010
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3M01
 
 | | The Crystal Structure of 5-epi-aristolochene synthase complexed with (2-trans,6-trans)-2-fluorofarnesyl diphosphate | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, ACETATE ION, Aristolochene synthase, ... | | Authors: | Noel, J.P, Dellas, N, Faraldos, J.A, Zhao, M, Hess Jr, B.A, Smentek, L, Coates, R.M, O'Maille, P.E. | | Deposit date: | 2010-03-02 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates.
Acs Chem.Biol., 5, 2010
|
|
7N6M
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RQKPLLGLSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N6K
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RALALLPLSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
