2GV5
 
 | | crystal structure of Sfi1p/Cdc31p complex | | Descriptor: | Cell division control protein 31, Sfi1p | | Authors: | Li, S, Sandercock, A.M, Conduit, P.T, Robinson, C.V, Williams, R.L, Kilmartin, J.V. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication.
J.Cell Biol., 173, 2006
|
|
3GXP
 
 | | Crystal structure of acid-alpha-galactosidase A complexed with galactose at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
2LT7
 
 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GXF
 
 | |
3GXT
 
 | |
2HBU
 
 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | Authors: | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
3IM1
 
 | |
3IT1
 
 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with L(+)-tartrate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, Acid phosphatase, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
3GK5
 
 | | Crystal structure of rhodanese-related protein (TVG0868615) from Thermoplasma volcanium, Northeast Structural Genomics Consortium Target TvR109A | | Descriptor: | Uncharacterized rhodanese-related protein TVG0868615 | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of rhodanese-related protein (TVG0868615) from Thermoplasma volcanium, Northeast Structural Genomics Consortium Target TvR109A
To be Published
|
|
3GR9
 
 | | Crystal structure of ColD H188K S187N | | Descriptor: | 2-OXOGLUTARIC ACID, ColD | | Authors: | Holden, H.M, Cook, P.D, Kubiak, R.L, Toomey, D.P. | | Deposit date: | 2009-03-25 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Site-Directed Mutations Are Required for the Conversion of a Sugar Dehydratase into an Aminotransferase.
Biochemistry, 48, 2009
|
|
3GXK
 
 | | The crystal structure of g-type lysozyme from Atlantic cod (Gadus morhua L.) in complex with NAG oligomers sheds new light on substrate binding and the catalytic mechanism. Native structure to 1.9 | | Descriptor: | COBALT (II) ION, Goose-type lysozyme 1 | | Authors: | Helland, R, Larsen, R.L, Finstad, S, Kyomuhendo, P, Larsen, A.N. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism.
Cell.Mol.Life Sci., 66, 2009
|
|
3GOZ
 
 | | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148 | | Descriptor: | Leucine-rich repeat-containing protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Belote, R.L, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-20 | | Release date: | 2009-03-31 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of the leucine-rich repeat-containing protein LegL7 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR148.
To be Published
|
|
2KT9
 
 | | Solution NMR Structure of Probable 30S Ribosomal Protein PSRP-3 (Ycf65-like protein) from Synechocystis sp. (strain PCC 6803), Northeast Structural Genomics Consortium Target Target SgR46 | | Descriptor: | Probable 30S ribosomal protein PSRP-3 | | Authors: | Liu, G, Janjua, J, Xiao, R, Mao, B, Buchwald, W.A, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Probable 30S ribosomal protein PSRP-3 (Ycf65-like protein) from Synechocystis sp. (PCC 6803), Northeast Structural Genomics Consortium Target Target SgR46
To be Published
|
|
2KTA
 
 | | Solution NMR structure of a domain of protein A6KY75 from Bacteroides vulgatus, Northeast Structural Genomics target BvR106A | | Descriptor: | Putative helicase | | Authors: | Mills, J.L, Sukumaran, D.K, Sathyamoorthy, B, Belote, R.L, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain of protein A6KY75 from Bacteroides vulgatus, Northeast Structural Genomics target BvR106A
To be Published
|
|
3GXN
 
 | | Crystal structure of apo alpha-galactosidase A at pH 4.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, SULFATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
2HBT
 
 | | Crystal structure of HIF prolyl hydroxylase EGLN-1 in complex with a biologically active inhibitor | | Descriptor: | Egl nine homolog 1, FE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE | | Authors: | Evdokimov, A.G, Walter, R.L, Mekel, M, Pokross, M.E, Kawamoto, R, Boyer, A. | | Deposit date: | 2006-06-14 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HIF prolyl hydroxylase in complex with a biologically active inhibitor
To be Published
|
|
2CHX
 
 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CAY
 
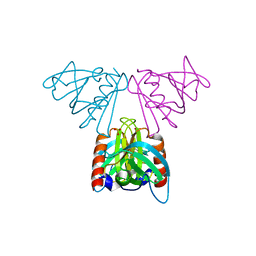 | | Vps36 N-terminal PH domain | | Descriptor: | SULFATE ION, VACUOLAR PROTEIN SORTING PROTEIN 36 | | Authors: | Teo, H, Williams, R.L, Perisic, O, Gill, D.J. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
2CAZ
 
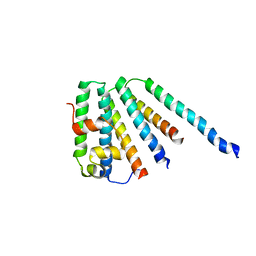 | | ESCRT-I core | | Descriptor: | PROTEIN SRN2, SUPPRESSOR PROTEIN STP22 OF TEMPERATURE-SENSITIVE ALPHA-FACTOR RECEPTOR AND ARGININE PERMEASE, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN VPS28 | | Authors: | Gill, D.J, Teo, H, Sun, J, Perisic, O, Veprintsev, D.B, Vallis, Y, Emr, S.D, Williams, R.L. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
2NQI
 
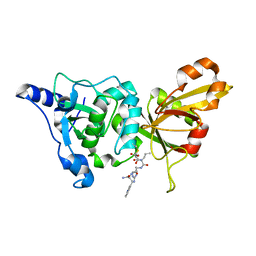 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
3IT0
 
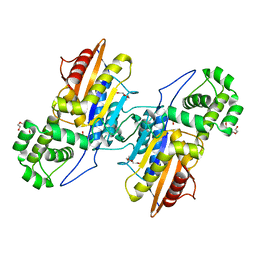 | | Crystal Structure Francisella tularensis histidine acid phosphatase complexed with phosphate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Acid phosphatase, PENTAETHYLENE GLYCOL, ... | | Authors: | Singh, H, Felts, R.L, Reilly, T.J, Tanner, J.J. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Crystal Structures of the histidine acid phosphatase from Francisella tularensis provide insight into substrate recognition.
J.Mol.Biol., 394, 2009
|
|
2GKO
 
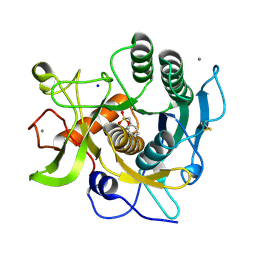 | | S41 Psychrophilic Protease | | Descriptor: | CALCIUM ION, SODIUM ION, microbial serine proteinases; subtilisin, ... | | Authors: | Walter, R.L, Mekel, M.J, Grayling, R.A, Arnold, F.H, Wintrode, P.L, Almog, O. | | Deposit date: | 2006-04-03 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
2GTN
 
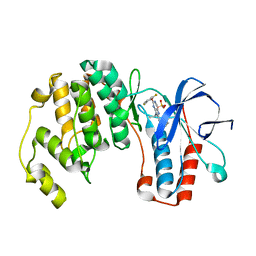 | | Mutated MAP kinase P38 (Mus Musculus) in complex with Inhbitor PG-951717 | | Descriptor: | 2-(2,6-DIFLUOROPHENOXY)-N-(2-FLUOROPHENYL)-9-ISOPROPYL-9H-PURIN-8-AMINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Sabat, M. | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The development of novel C-2, C-8, and N-9 trisubstituted purines as inhibitors of TNF-alpha production.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2CHW
 
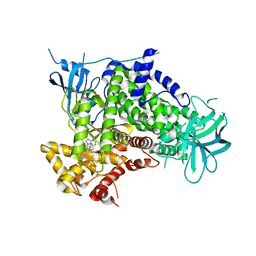 | | A pharmacological map of the PI3-K family defines a role for p110 alpha in signaling: The structure of complex of phosphoinositide 3- kinase gamma with inhibitor PIK-39 | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2NQG
 
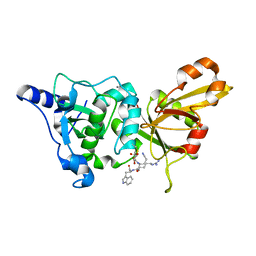 | | Calpain 1 proteolytic core inactivated by WR18(S,S), an epoxysuccinyl-type inhibitor. | | Descriptor: | 5-AZANYLIDYNE-N-[(2S)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]-L-NORVALYL-L-ARGINYL-L-TRYPTOPHANAMIDE, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
