1KIR
 
 | |
7WZO
 
 | |
7XXK
 
 | | Crystal structure of SARS-CoV-2 N-CTD in complex with GMP | | Descriptor: | CHLORIDE ION, GUANINE, GUANOSINE, ... | | Authors: | Zhou, R.J, Ni, X.C, Lei, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the C-terminal domain of SARS-CoV-2 N protein in complex with GMP reveals critical residues for RNA interaction.
Bioorg.Med.Chem.Lett., 2024
|
|
1UMX
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ARG M267 REPLACED WITH LEU (CHAIN M, R267L) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, FE (III) ION, ... | | Authors: | Fyfe, P.K, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2003-09-02 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre.
Biochim.Biophys.Acta, 1608, 2004
|
|
1B2M
 
 | | THREE-DIMENSIONAL STRUCTURE OF RIBONULCEASE T1 COMPLEXED WITH AN ISOSTERIC PHOSPHONATE ANALOGUE OF GPU: ALTERNATE SUBSTRATE BINDING MODES AND CATALYSIS. | | Descriptor: | 5'-R(*GP*(U34))-3', RIBONUCLEASE T1 | | Authors: | Arni, R.K, Watanabe, L, Ward, R.J, Kreitman, R.J, Kumar, K, Walz Jr, F.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with an isosteric phosphonate substrate analogue of GpU: alternate substrate binding modes and catalysis.
Biochemistry, 38, 1999
|
|
7YX8
 
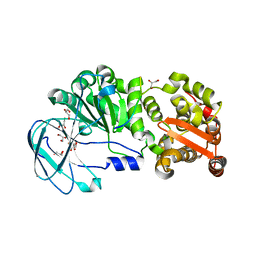 | | Crystal structure of the AM0627 (E326A) inactive mutant in complex with PSGL-1-like bis-T glycopeptide and Zn2+ | | Descriptor: | GLYCEROL, PSGL-1-like bis-T glycopeptide, Peptidase M60 domain-containing protein, ... | | Authors: | Taleb, V, Liao, Q, Narimatsu, Y, Garcia-Garcia, A, Companon, I, Borges, R.J, Gonzalez-Ramirez, A.M, Corzana, F, Clausen, H, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the cleavage of clustered O-glycan patches-containing glycoproteins by mucinases of the human gut.
Nat Commun, 13, 2022
|
|
7YX3
 
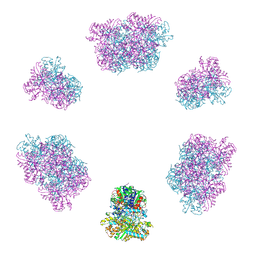 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
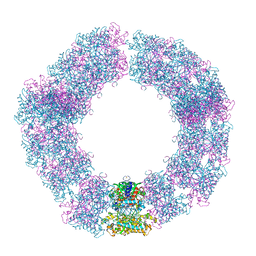 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
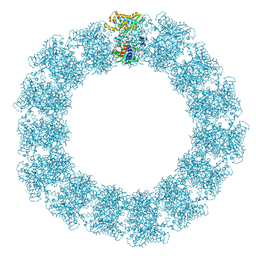 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
1KIP
 
 | |
1KIQ
 
 | |
7YYK
 
 | | Crystal structure of the O-fucosylated form of TSRs1-3 from the human thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1, alpha-L-fucopyranose | | Authors: | Berardinelli, S.J, Eletsky, A, Valero-Gonzalez, J, Ito, A, Manjunath, R, Hurtado-Guerrero, R, Prestegard, J.R, Woods, R.J, Haltiwanger, R.S. | | Deposit date: | 2022-02-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | O-fucosylation stabilizes the TSR3 motif in thrombospondin-1 by interacting with nearby amino acids and protecting a disulfide bond.
J.Biol.Chem., 298, 2022
|
|
1JR5
 
 | | Solution Structure of the Anti-Sigma Factor AsiA Homodimer | | Descriptor: | 10 KDA Anti-Sigma Factor | | Authors: | Urbauer, J.L, Simeonov, M.F, Bieber Urbauer, R.J, Adelman, K, Gilmore, J.M, Brody, E.N. | | Deposit date: | 2001-08-10 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3Q9O
 
 | |
1MG1
 
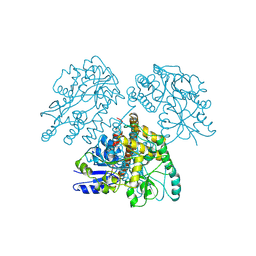 | | HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA | | Descriptor: | CHLORIDE ION, PROTEIN (HTLV-1 GP21 ECTODOMAIN/MALTOSE-BINDING PROTEIN CHIMERA), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kobe, B, Center, R.J, Kemp, B.E, Poumbourios, P. | | Deposit date: | 1999-03-01 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2F33
 
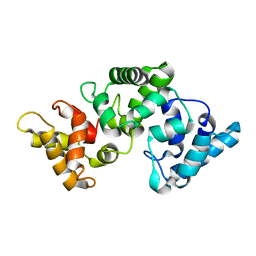 | | NMR solution structure of Ca2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FFH
 
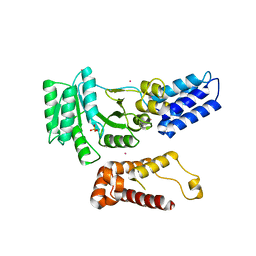 | | THE SIGNAL SEQUENCE BINDING PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | CADMIUM ION, PROTEIN (FFH), SULFATE ION | | Authors: | Keenan, R.J, Freymann, D.M, Walter, P, Stroud, R.M. | | Deposit date: | 1999-06-29 | | Release date: | 1999-07-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the signal sequence binding subunit of the signal recognition particle.
Cell(Cambridge,Mass.), 94, 1998
|
|
2G9B
 
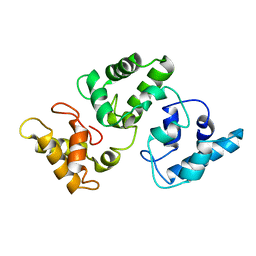 | | NMR solution structure of CA2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2006-03-06 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
4PKB
 
 | |
7ZDI
 
 | | PucB-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3D1O
 
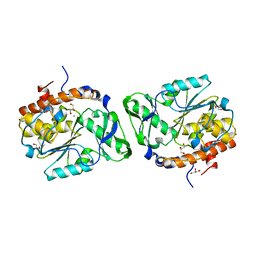 | | Structure of the PTP-Like Phytase Expressed by Selenomonas Ruminantium at an Ionic Strength of 300 mM | | Descriptor: | CHLORIDE ION, GLYCEROL, Myo-inositol hexaphosphate phosphohydrolase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effect of ionic strength and oxidation on the P-loop conformation of the protein tyrosine phosphatase-like phytase, PhyAsr.
Febs J., 275, 2008
|
|
7ZE3
 
 | | PucD-LH2 complex from Rps. palustris | | Descriptor: | (3'E)-3',4'-didehydro-1,2-dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZE8
 
 | | PucE-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein, ... | | Authors: | Qian, P, Cogdell, R.J, Nguyen-Phan, T.C. | | Deposit date: | 2022-03-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3CKZ
 
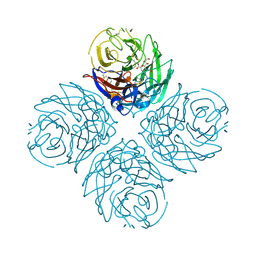 | | N1 Neuraminidase H274Y + Zanamivir | | Descriptor: | CALCIUM ION, Neuraminidase, ZANAMIVIR | | Authors: | Colllins, P, Haire, L.F, Lin, Y.P, Liu, J, Russell, R.J, Walker, P.A, Skehel, J.J, Martin, S.R, Hay, A.J, Gamblin, S.J. | | Deposit date: | 2008-03-18 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants.
Nature, 453, 2008
|
|
7ZCU
 
 | | PucA-LH2 complex from Rps. palustris | | Descriptor: | 1,2-Dihydro-psi,psi-caroten-1-ol, BACTERIOCHLOROPHYLL A, Light-harvesting protein B-800-850 alpha chain, ... | | Authors: | Qian, P, Cogdell, R.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of light-harvesting 2 complexes from Rhodopseudomonas palustris reveal the molecular origin of absorption tuning.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
