2CNZ
 
 | | Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide (V13A mutant) | | Descriptor: | PUTATIVE OUTER MEMBRANE PROTEIN | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
2CO2
 
 | | Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide (F3A mutant) | | Descriptor: | SAFA N-TERMINAL EXTENSION, SAFA PILUS SUBUNIT | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
2CNY
 
 | | Salmonella enterica SafA pilin in complex with a 19-residue SafA Nte peptide (I15A mutant) | | Descriptor: | PUTATIVE OUTER MEMBRANE PROTEIN | | Authors: | Remaut, H, Rose, R.J, Hannan, T.J, Hultgren, S.J, Radford, S.E, Ashcroft, A.E, Waksman, G. | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Donor-Strand Exchange in Chaperone-Assisted Pilus Assembly Proceeds Through a Concerted Beta-Strand Displacement Mechanism
Mol.Cell, 22, 2006
|
|
3TJJ
 
 | | Crystal structure of human peroxiredoxin IV C245A mutant in sulfenylated form | | Descriptor: | Peroxiredoxin-4 | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TJF
 
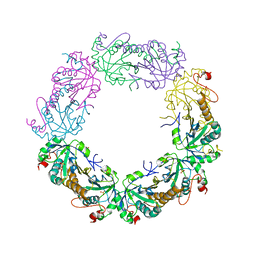 | | Crystal Structure of human peroxiredoxin IV C51A mutant in reduced form | | Descriptor: | Peroxiredoxin-4, SULFATE ION | | Authors: | Cao, Z, Tavender, T.J, Roszak, A.W, Cogdell, R.J, Bulleid, N.J. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of Reduced and of Oxidized Peroxiredoxin IV Enzyme Reveals a Stable Oxidized Decamer and a Non-disulfide-bonded Intermediate in the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
5CQO
 
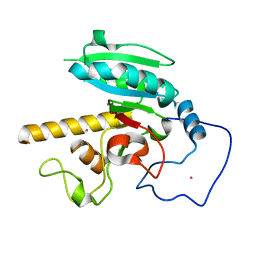 | | GTB mutant with mercury - E303D | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-21 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
5CQL
 
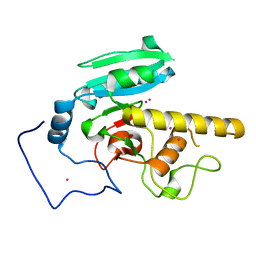 | | GTB mutant with mercury - E303A | | Descriptor: | Histo-blood group ABO system transferase, MERCURY (II) ION | | Authors: | Gagnon, S.M.L, Blackler, R.J. | | Deposit date: | 2015-07-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Glycosyltransfer in mutants of putative catalytic residue Glu303 of the human ABO(H) A and B blood group glycosyltransferases GTA and GTB proceeds through a labile active site.
Glycobiology, 27, 2017
|
|
2CHO
 
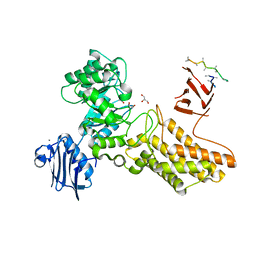 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
5CXU
 
 | |
2CEO
 
 | | thyroxine-binding globulin complex with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, GLYCEROL, THYROXINE-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J, Carrell, R.W. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Mechanism for the Carriage and Release of Thyroxine in the Blood.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3T6E
 
 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-94 | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
2F4U
 
 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-((1R,2S,3R,4R,5S)-5-AMINO-4-[(2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-{2-[(3-AMINOPROPYL)AMINO]ETHOXY}-3-HYDROXYCYCLOHEXYL)-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2F4V
 
 | | 30S ribosome + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, (2S,4S,4AR,5AS,6S,11R,11AS,12R,12AR)-7-CHLORO-4-(DIMETHYLAMINO)-6,10,11,12-TETRAHYDROXY-1,3-DIOXO-1,2,3,4,4A,5,5A,6,11,11A,12,12A-DODECAHYDROTETRACENE-2-CARBOXAMIDE, 16S ribosomal RNA, ... | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosome aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
5BTA
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
5BTC
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2F13
 
 | | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor | | Descriptor: | (2R)-2,3-dihydroxypropyl 2-acetamido-2,4-dideoxy-alpha-L-threo-hex-4-enopyranosiduronic acid, PHOSPHATE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Kato, R, Mann, M.C, Thomson, R.J, Dyason, J.C, von Itzstein, M, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Monti, E, Wakatsuki, S. | | Deposit date: | 2005-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor
To be Published
|
|
3TX8
 
 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U6Q
 
 | | MutM set 2 ApGo | | Descriptor: | DNA (5'-D(*AP*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*(CX2)P*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U6E
 
 | | MutM set 1 TpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*(8OG)P*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
5BTF
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(3S)-3-methylpiperazin-1-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BS8
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BTI
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | (3S)-9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6V9D
 
 | |
5BTD
 
 | | Crystal structure of a topoisomerase II complex | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(3S)-3-methylpiperazin-1-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Blower, T.R, Williamson, B.H, Kerns, R.J, Berger, J.M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal structure and stability of gyrase-fluoroquinolone cleaved complexes from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
