1ANC
 
 | |
1A98
 
 | | XPRTASE FROM E. COLI COMPLEXED WITH GMP | | Descriptor: | XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1AND
 
 | |
1ANB
 
 | |
1A96
 
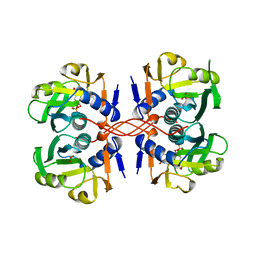 | | XPRTASE FROM E. COLI WITH BOUND CPRPP AND XANTHINE | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Vos, S, Parry, R.J, Burns, M.R, De Jersey, J, Martin, J.L. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of free and complexed forms of Escherichia coli xanthine-guanine phosphoribosyltransferase.
J.Mol.Biol., 282, 1998
|
|
1EVU
 
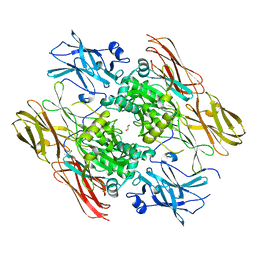 | | HUMAN FACTOR XIII WITH CALCIUM BOUND IN THE ION SITE | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII, S-1,2-PROPANEDIOL | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-20 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To Be Published
|
|
1EX0
 
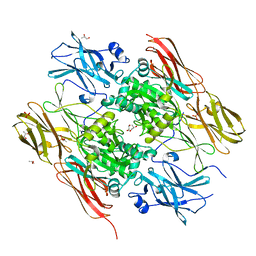 | | HUMAN FACTOR XIII, MUTANT W279F ZYMOGEN | | Descriptor: | CALCIUM ION, COAGULATION FACTOR XIII A CHAIN, PHOSPHATE ION, ... | | Authors: | Garzon, R.J, Pratt, K.P, Bishop, P.D, Le Trong, I, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 2000-04-28 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tryptophan 279 is Essential for the Transglutaminase Activity of Coagulation Factor XIII: Functional and Structural Characterization
To be Published
|
|
1AV3
 
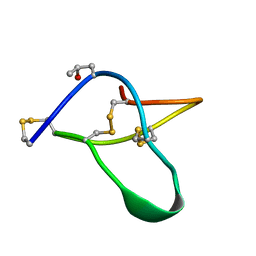 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1EFY
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC FRAGMENT OF POLY (ADP-RIBOSE) POLYMERASE COMPLEXED WITH A BENZIMIDAZOLE INHIBITOR | | Descriptor: | 2-(3'-METHOXYPHENYL) BENZIMIDAZOLE-4-CARBOXAMIDE, POLY (ADP-RIBOSE) POLYMERASE | | Authors: | White, A.W, Almassy, R, Calvert, A.H, Curtin, N.J, Griffin, R.J, Hostomsky, Z, Maegley, K, Newell, D.R, Srinivasan, S, Golding, B.T. | | Deposit date: | 2000-02-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase.
J.Med.Chem., 43, 2000
|
|
1B8W
 
 | | DEFENSIN-LIKE PEPTIDE 1 | | Descriptor: | PROTEIN (DEFENSIN-LIKE PEPTIDE 1) | | Authors: | Torres, A.M, Wang, X, Fletcher, J.I, Alewood, D, Alewood, P.F, Smith, R, Simpson, R.J, Nicholson, G.M, Sutherland, S.K, Gallagher, C.H, King, G.F, Kuchel, P.W. | | Deposit date: | 1999-02-02 | | Release date: | 1999-09-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a defensin-like peptide from platypus venom.
Biochem.J., 341, 1999
|
|
1BG2
 
 | | HUMAN UBIQUITOUS KINESIN MOTOR DOMAIN | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, KINESIN, ... | | Authors: | Kull, F.J, Sablin, E.P, Lau, R, Fletterick, R.J, Vale, R.D. | | Deposit date: | 1998-06-04 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the kinesin motor domain reveals a structural similarity to myosin.
Nature, 380, 1996
|
|
1EJ8
 
 | | CRYSTAL STRUCTURE OF DOMAIN 2 OF THE YEAST COPPER CHAPERONE FOR SUPEROXIDE DISMUTASE (LYS7) AT 1.55 A RESOLUTION | | Descriptor: | CALCIUM ION, LYS7 | | Authors: | Hall, L.T, Sanchez, R.J, Holloway, S.P, Zhu, H, Stine, J.E, Lyons, T.J, Demeler, B, Schirf, V, Hansen, J.C, Nersissian, A.M, Valentine, J.S, Hart, P.J. | | Deposit date: | 2000-03-01 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic and analytical ultracentrifugation analyses of truncated and full-length yeast copper chaperones for SOD (LYS7): a dimer-dimer model of LYS7-SOD association and copper delivery.
Biochemistry, 39, 2000
|
|
1AP4
 
 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-SATURATED STATE, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, CARDIAC N-TROPONIN C | | Authors: | Li, M.X, Spyracopoulos, L, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1EZU
 
 | | ECOTIN Y69F, D70P BOUND TO D102N TRYPSIN | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZS
 
 | | CRYSTAL STRUCTURE OF ECOTIN MUTANT M84R, W67A, G68A, Y69A, D70A BOUND TO RAT ANIONIC TRYPSIN II | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZE
 
 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
1BPJ
 
 | | THYMIDYLATE SYNTHASE R178T, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BY6
 
 | |
1FLJ
 
 | | CRYSTAL STRUCTURE OF S-GLUTATHIOLATED CARBONIC ANHYDRASE III | | Descriptor: | CARBONIC ANHYDRASE III, GLUTATHIONE, ZINC ION | | Authors: | Mallis, R.J, Poland, B.W, Chatterjee, T.K, Fisher, R.A, Darmawan, S, Honzatko, R.B, Thomas, J.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of S-glutathiolated carbonic anhydrase III.
FEBS Lett., 482, 2000
|
|
1DZO
 
 | | Truncated PAK pilin from Pseudomonas aeruginosa | | Descriptor: | TYPE IV PILIN | | Authors: | Hazes, B, Read, R.J. | | Deposit date: | 2000-03-06 | | Release date: | 2000-06-11 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of Pseudomonas Aeruginosa Pak Pilin Suggests a Main-Chain-Dominated Mode of Receptor Binding
J.Mol.Biol., 299, 2000
|
|
1E3A
 
 | | A slow processing precursor penicillin acylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hewitt, L, Kasche, V, Lummer, K, Lewis, R.J, Murshudov, G.N, Verma, C.S, Dodson, G.G, Wilson, K.S. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Slow Processing Precursor Penicillin Acylase from Escherichia Coli Reveals the Linker Peptide Blocking the Active-Site Cleft
J.Mol.Biol., 302, 2000
|
|
1DTJ
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | RNA-BINDING NEUROONCOLOGICAL VENTRAL ANTIGEN 2 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L, Musunuru, K, Zhong, R, Darnell, R.B, Burley, S.K. | | Deposit date: | 2000-01-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1B3K
 
 | | Plasminogen activator inhibitor-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Sharp, A.M, Stein, P.E, Pannu, N.S, Read, R.J. | | Deposit date: | 1998-12-11 | | Release date: | 1999-12-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion.
Structure Fold.Des., 7, 1999
|
|
1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
