2WJW
 
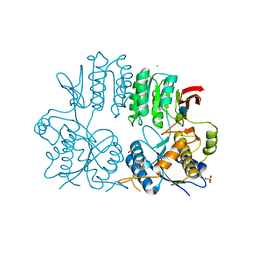 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 1.8 A resolution | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | 著者 | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | 登録日 | 2009-06-01 | | 公開日 | 2009-08-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
2VW1
 
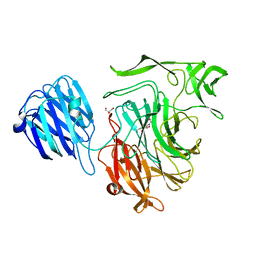 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | 分子名称: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, SIALIDASE B | | 著者 | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | 登録日 | 2008-06-13 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
3NTR
 
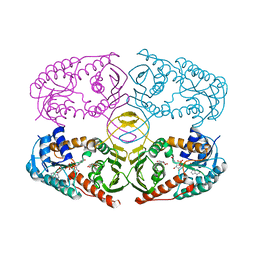 | | Crystal structure of K97V mutant of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NAD and inositol | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-07-05 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.6503 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
4C4N
 
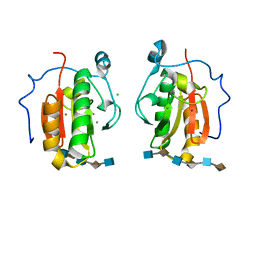 | | Crystal structure of the Sonic Hedgehog-heparin complex | | 分子名称: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, CALCIUM ION, ... | | 著者 | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | 登録日 | 2013-09-05 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4C4M
 
 | | Crystal structure of the Sonic Hedgehog-chondroitin-4-sulphate complex | | 分子名称: | ACETATE ION, CALCIUM ION, SONIC HEDGEHOG PROTEIN, ... | | 著者 | Whalen, D.M, Malinauskas, T, Gilbert, R.J.C, Siebold, C. | | 登録日 | 2013-09-05 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural Insights Into Proteoglycan-Shaped Hedgehog Signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4E7X
 
 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | 分子名称: | ACETATE ION, Poly(A) RNA polymerase protein cid1 | | 著者 | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | 登録日 | 2012-03-19 | | 公開日 | 2012-07-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3O9Q
 
 | | Effector domain of NS1 from A/PR/8/34 containing a W187A mutation | | 分子名称: | Nonstructural protein 1 | | 著者 | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | 登録日 | 2010-08-04 | | 公開日 | 2011-05-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9T
 
 | | Effector domain from influenza A/PR/8/34 NS1 | | 分子名称: | HEXAETHYLENE GLYCOL, Nonstructural protein 1 | | 著者 | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | 登録日 | 2010-08-04 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
3O9U
 
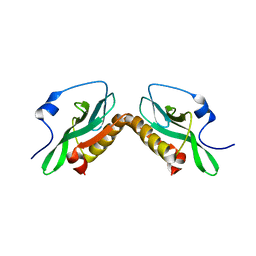 | | Effector domain of influenza A/PR/8/34 NS1 | | 分子名称: | Nonstructural protein 1 | | 著者 | Kerry, P.S, Lewis, A, Hale, B.G, Hass, C, Taylor, M.A, Randall, R.E, Russell, R.J.M. | | 登録日 | 2010-08-04 | | 公開日 | 2011-05-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | A Transient Homotypic Interaction Model for the Influenza A Virus NS1 Protein Effector Domain.
Plos One, 6, 2011
|
|
2X44
 
 | | Structure of a strand-swapped dimeric form of CTLA-4 | | 分子名称: | CYTOTOXIC T-LYMPHOCYTE PROTEIN 4 | | 著者 | Sonnen, A.F.-P, Yu, C, Evans, E.J, Stuart, D.I, Davis, S.J, Gilbert, R.J.C. | | 登録日 | 2010-01-28 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Domain Metastability: A Molecular Basis for Immunoglobulin Deposition?
J.Mol.Biol., 399, 2010
|
|
2WJX
 
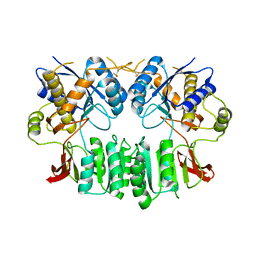 | | Crystal structure of the human ionotropic glutamate receptor GluR2 ATD region at 4.1 A resolution | | 分子名称: | GLUTAMATE RECEPTOR 2 | | 著者 | Clayton, A, Siebold, C, Gilbert, R.J.C, Sutton, G.C, Harlos, K, McIlhinney, R.A.J, Jones, E.Y, Aricescu, A.R. | | 登録日 | 2009-06-01 | | 公開日 | 2009-08-18 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Crystal Structure of the Glur2 Amino-Terminal Domain Provides Insights Into the Architecture and Assembly of Ionotropic Glutamate Receptors.
J.Mol.Biol., 392, 2009
|
|
3MZ0
 
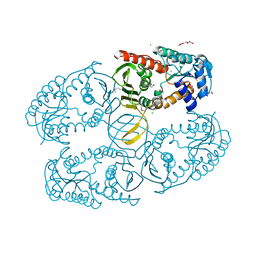 | | Crystal structure of apo myo-inositol dehydrogenase from Bacillus subtilis | | 分子名称: | CHLORIDE ION, GLYCEROL, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2010-05-11 | | 公開日 | 2010-09-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
2XQ5
 
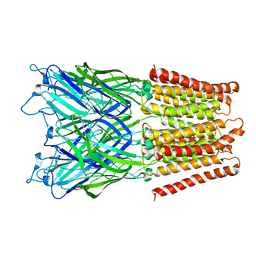 | | Pentameric ligand gated ion channel GLIC in complex with tetraethylarsonium (TEAs) | | 分子名称: | ARSENIC, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ4
 
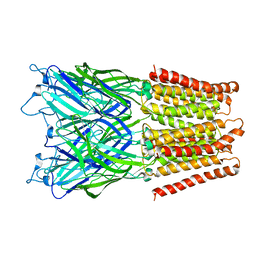 | | Pentameric ligand gated ion channel GLIC in complex with tetramethylarsonium (TMAs) | | 分子名称: | ARSENIC, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ9
 
 | | Pentameric ligand gated ion channel GLIC mutant E221A in complex with tetraethylarsonium (TEAs) | | 分子名称: | ARSENIC, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ8
 
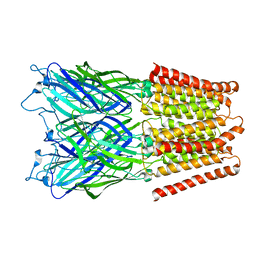 | | Pentameric ligand gated ion channel GLIC in complex with zinc ion (Zn2+) | | 分子名称: | GLR4197 PROTEIN, ZINC ION | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
1TL7
 
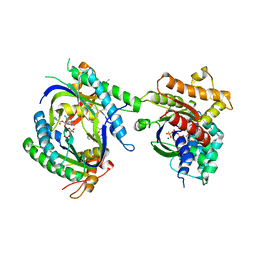 | | Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With 2'(3')-O-(N-methylanthraniloyl)-guanosine 5'-triphosphate and Mn | | 分子名称: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | 著者 | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | 登録日 | 2004-06-09 | | 公開日 | 2004-12-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
2XQ6
 
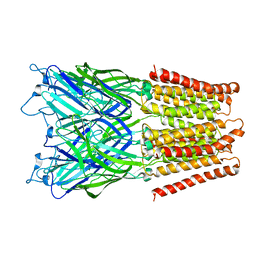 | | Pentameric ligand gated ion channel GLIC in complex with cesium ion (Cs+) | | 分子名称: | CESIUM ION, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQA
 
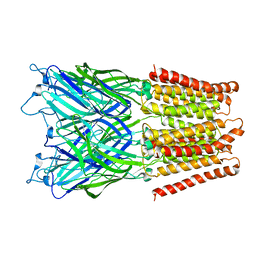 | | Pentameric ligand gated ion channel GLIC in complex with tetrabutylantimony (TBSb) | | 分子名称: | ANTIMONY (III) ION, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ7
 
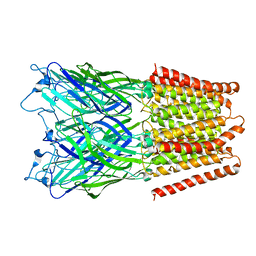 | | Pentameric ligand gated ion channel GLIC in complex with cadmium ion (Cd2+) | | 分子名称: | CADMIUM ION, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQ3
 
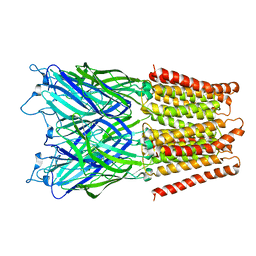 | | Pentameric ligand gated ion channel GLIC in complex with Br-lidocaine | | 分子名称: | BROMIDE ION, GLR4197 PROTEIN | | 著者 | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | 登録日 | 2010-09-01 | | 公開日 | 2010-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
3OSK
 
 | | Crystal structure of human CTLA-4 apo homodimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | 著者 | Yu, C, Sonnen, A.F.-P, Ikemizu, S, Stuart, D.I, Gilbert, R.J.C, Davis, S.J. | | 登録日 | 2010-09-09 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rigid-body ligand recognition drives cytotoxic T-lymphocyte antigen 4 (CTLA-4) receptor triggering
J.Biol.Chem., 286, 2011
|
|
4KS1
 
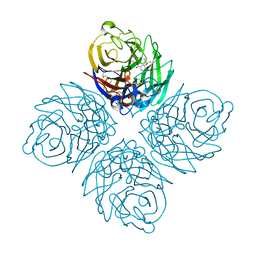 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | 分子名称: | (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Kerry, P.S, Russell, R.J.M. | | 登録日 | 2013-05-17 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
4K2B
 
 | |
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | 著者 | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | 登録日 | 2013-04-09 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
