1B9P
 
 | |
1JF8
 
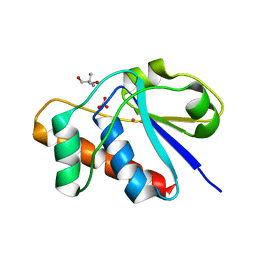 | | X-ray structure of reduced C10S, C15A arsenate reductase from pI258 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, POTASSIUM ION, ... | | Authors: | Zegers, I, Martins, J.C, Willem, R, Wyns, L, Messens, J. | | Deposit date: | 2001-06-20 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Arsenate reductase from S. aureus plasmid pI258 is a phosphatase drafted for redox duty.
Nat.Struct.Biol., 8, 2001
|
|
4CQM
 
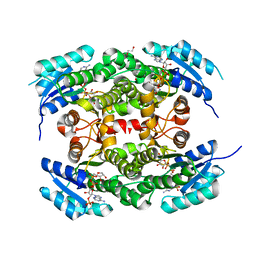 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD and NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBONYL REDUCTASE FAMILY MEMBER 4, ... | | Authors: | Venkatesan, R, SahTeli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastoniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
2HEH
 
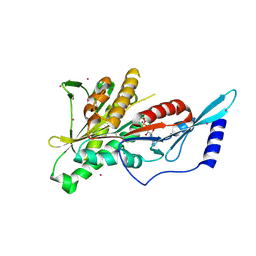 | | Crystal Structure of the KIF2C motor domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF2C protein, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the kif2c motor domain
to be published
|
|
1BHF
 
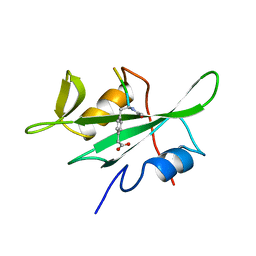 | | P56LCK SH2 DOMAIN INHIBITOR COMPLEX | | Descriptor: | INHIBITOR ACE-IPA-GLU-GLU-ILE, T-LYMPHOCYTE-SPECIFIC PROTEIN TYROSINE KINASE P56LCK | | Authors: | Tong, L, Warren, T.C, Lukas, S, Schembri-King, J, Betageri, R, Proudfoot, J.R, Jakes, S. | | Deposit date: | 1998-06-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carboxymethyl-phenylalanine as a replacement for phosphotyrosine in SH2 domain binding.
J.Biol.Chem., 273, 1998
|
|
4CHK
 
 | | Crystal Structure of the ARF5 oligomerization domain | | Descriptor: | AUXIN RESPONSE FACTOR 5 | | Authors: | Nanao, M.H, Mazzoleni, M, Thevenon, E, Brunoud, G, Vernoux, T, Parcy, F, Dumas, R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for Oligomerisation of Auxin Transcriptional Regulators
Nat.Commun., 5, 2014
|
|
2ZBY
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) | | Descriptor: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugimoto, H, Shinkyo, R, Hayashi, K, Yoneda, S, Yamada, M, Kamakura, M, Ikushiro, S, Shiro, Y, Sakaki, T. | | Deposit date: | 2007-10-30 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of CYP105A1 (P450SU-1) in Complex with 1alpha,25-Dihydroxyvitamin D3
Biochemistry, 47, 2008
|
|
1B9C
 
 | |
1BEO
 
 | | BETA-CRYPTOGEIN | | Descriptor: | BETA-CRYPTOGEIN | | Authors: | Boissy, G, De La Fortelle, E, Kahn, R, Huet, J.C, Bricogne, G, Pernollet, J.C, Brunie, S. | | Deposit date: | 1996-08-02 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a fungal elicitor secreted by Phytophthora cryptogea, a member of a novel class of plant necrotic proteins.
Structure, 4, 1996
|
|
2HC5
 
 | | Solution NMR Structure of Protein yvyC from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR482. | | Descriptor: | Hypothetical protein yvyC | | Authors: | Eletsky, A, Liu, G, Atreya, H.S, Sukumaran, D.K, Wang, D, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YvyC from Bacillus subtilis reveals unexpected structural similarity between two PFAM families.
Proteins, 76, 2009
|
|
1J9Z
 
 | | CYPOR-W677G | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
2ZF5
 
 | | Crystal Structure of highly thermostable glycerol kinase from a hyperthermophilic archaeon | | Descriptor: | Glycerol kinase | | Authors: | Koga, Y, Katsumi, R, You, D.-J, Matsumura, H, Takano, K, Kanaya, S. | | Deposit date: | 2007-12-20 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of highly thermostable glycerol kinase from a hyperthermophilic archaeon in a dimeric form
Febs J., 275, 2008
|
|
1JCO
 
 | | Solution structure of the monomeric [Thr(B27)->Pro,Pro(B28)->Thr] insulin mutant (PT insulin) | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Keller, D, Clausen, R, Josefsen, K, Led, J.J. | | Deposit date: | 2001-06-11 | | Release date: | 2001-10-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity.
Biochemistry, 40, 2001
|
|
3SR6
 
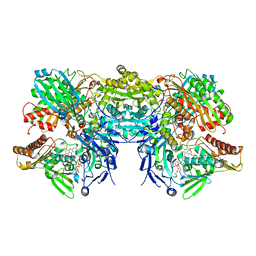 | | Crystal Structure of Reduced Bovine Xanthine Oxidase in Complex with Arsenite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Crystal Structure of Arsenite-Inhibited Xanthine Oxidase: Mu-Sulfido,Mu-Oxo Double Bridge between Molybdenum and Arsenic in the Active Site.
J.Am.Chem.Soc., 133, 2011
|
|
3SSC
 
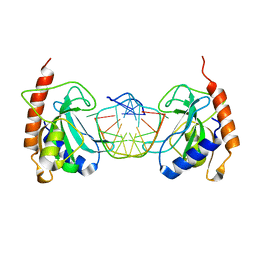 | | DNA binding domain of restriction endonuclease bound to DNA | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, DNA (5'-D(*AP*GP*CP*TP*AP*(5CM)P*CP*GP*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*(5CM)P*CP*GP*GP*TP*AP*GP*C)-3') | | Authors: | Sukackaite, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2011-07-08 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine.
Nucleic Acids Res., 40, 2012
|
|
4AKL
 
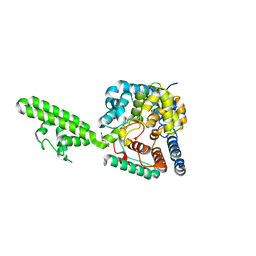 | | Structure of the Crimean-Congo Haemorrhagic Fever Virus Nucleocapsid Protein | | Descriptor: | NUCLEOCAPSID, TRIETHYLENE GLYCOL | | Authors: | Carter, S.D, Walter, C.T, Surtees, R, Bergeron, E, Ariza, A, Albarino, C.G, Nichol, S.T, Hiscox, J.A, Edwards, T.A, Barr, J.N. | | Deposit date: | 2012-02-24 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, Function, and Evolution of the Crimean-Congo Hemorrhagic Fever Virus Nucleocapsid Protein.
J.Virol., 86, 2012
|
|
1JEO
 
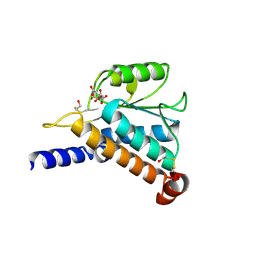 | | Crystal Structure of the Hypothetical Protein MJ1247 from Methanococcus jannaschii at 2.0 A Resolution Infers a Molecular Function of 3-Hexulose-6-Phosphate isomerase. | | Descriptor: | CITRIC ACID, HYPOTHETICAL PROTEIN MJ1247 | | Authors: | Martinez-Cruz, L.A, Dreyer, M.K, Boisvert, D.C, Yokota, H, Martinez-Chantar, M.L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2001-06-18 | | Release date: | 2002-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MJ1247 protein from M. jannaschii at 2.0 A resolution infers a molecular function of 3-hexulose-6-phosphate isomerase.
Structure, 10, 2002
|
|
4AMU
 
 | | Structure of ornithine carbamoyltransferase from Mycoplasma penetrans with a P321 space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
462D
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 GENOMIC RNA DIMERIZATION INITIATION SITE | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*UP*UP*GP*CP*UP*GP*AP*GP*GP*UP*GP*CP*AP*CP*AP*CP*AP*GP*CP*AP*AP*G) -3') | | Authors: | Ennifar, E, Yusupov, M, Walter, P, Marquet, R, Ehresmann, C, Ehresmann, B, Dumas, P. | | Deposit date: | 1999-03-18 | | Release date: | 1999-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges.
Structure Fold.Des., 7, 1999
|
|
4A22
 
 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to N-(4-hydroxybutyl)- glycolohydroxamic acid bis- phosphate | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, ... | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli.
J. Biol. Chem., 286, 2011
|
|
5R4O
 
 | | PanDDA analysis group deposition of ground-state model of BROMODOMAIN OF HUMAN NUCLEOSOME-REMODELING FACTOR SUBUNIT BPTF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, Nucleosome-remodeling factor subunit BPTF | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Renjie, Z, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
2ZHM
 
 | | Crystal structure of human galectin-9 N-terminal CRD in complex with N-acetyllactosamine trimer (crystal 1) | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, Galectin-9, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nagae, M, Nishi, N, Murata, T, Usui, T, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2008-02-06 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain.
Glycobiology, 19, 2009
|
|
2ZJ2
 
 | | Archaeal DNA helicase Hjm apo state in form 1 | | Descriptor: | Putative ski2-type helicase, SULFATE ION | | Authors: | Oyama, T, Oka, H, Fujikane, R, Ishino, Y, Morikawa, K. | | Deposit date: | 2008-02-29 | | Release date: | 2009-02-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm
Bmc Struct.Biol., 9, 2009
|
|
1B8W
 
 | | DEFENSIN-LIKE PEPTIDE 1 | | Descriptor: | PROTEIN (DEFENSIN-LIKE PEPTIDE 1) | | Authors: | Torres, A.M, Wang, X, Fletcher, J.I, Alewood, D, Alewood, P.F, Smith, R, Simpson, R.J, Nicholson, G.M, Sutherland, S.K, Gallagher, C.H, King, G.F, Kuchel, P.W. | | Deposit date: | 1999-02-02 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a defensin-like peptide from platypus venom.
Biochem.J., 341, 1999
|
|
2ZVO
 
 | | NEMO CoZi domain in complex with diubiquitin in C2 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
