3WY9
 
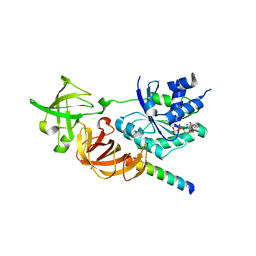 | | Crystal structure of a complex of the archaeal ribosomal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1alpha | | Descriptor: | 50S ribosomal protein L12, Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
6K0L
 
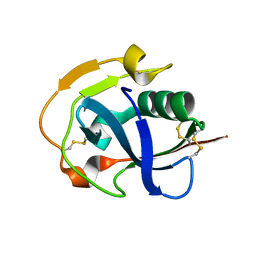 | | The crystal structure of simian CD163 SRCR5 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
1SAU
 
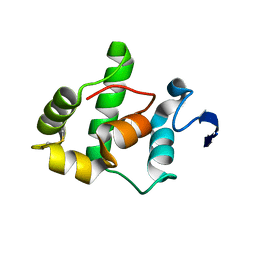 | | The Gamma subunit of the dissimilatory sulfite reductase (DsrC) from Archaeoglobus fulgidus at 1.1 A resolution | | Descriptor: | sulfite reductase, desulfoviridin-type subunit gamma | | Authors: | Mander, G.J, Weiss, M.S, Hedderich, R, Kahnt, J, Ermler, U, Warkentin, E. | | Deposit date: | 2004-02-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Determination of a novel structure by a combination of long-wavelength sulfur phasing and radiation-damage-induced phasing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6K73
 
 | |
4L78
 
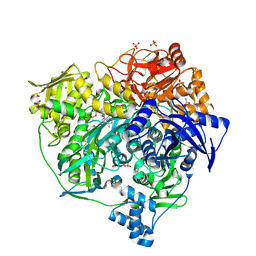 | | Xenon Trapping and Statistical Coupling Analysis Uncover Regions Important for Structure and Function of Multidomain Protein StPurL | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tanwar, A.S, Goyal, V.D, Choudhary, D, Panjikar, S, Anand, R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Importance of hydrophobic cavities in allosteric regulation of formylglycinamide synthetase: insight from xenon trapping and statistical coupling analysis
Plos One, 8, 2013
|
|
3ZCU
 
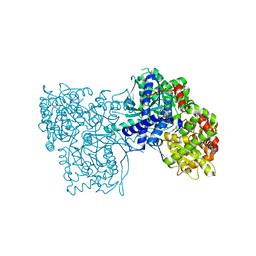 | | Rabbit muscle glycogen phosphorylase b in complex with N-(pyridyl-2- carbonyl)-N-beta-D-glucopyranosyl urea determined at 2.05 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(pyridin-2-ylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
7EVA
 
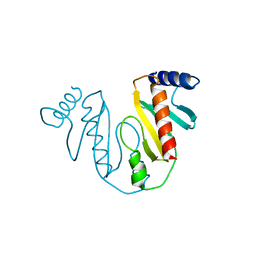 | |
6K6W
 
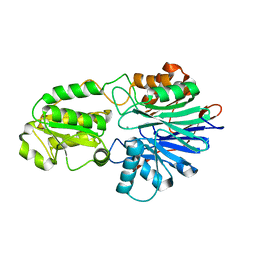 | |
3ZLP
 
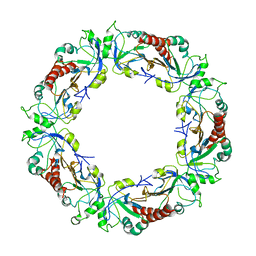 | | Crystal structure of Schistosoma mansoni Peroxiredoxin 1 C48P mutant form with four decamers in the asymmetric unit | | Descriptor: | THIOREDOXIN PEROXIDASE | | Authors: | Saccoccia, F, Angelucci, F, Ardini, M, Boumis, G, Brunori, M, DiLeandro, L, Ippoliti, R, Miele, A.E, Natoli, G, Scotti, S, Bellelli, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.515 Å) | | Cite: | Switching between the Alternative Structures and Functions of a 2-Cys Peroxiredoxin, by Site-Directed Mutagenesis
J.Mol.Biol., 425, 2013
|
|
6I7Y
 
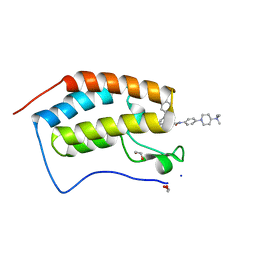 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [1-[4-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]phenyl]piperidin-4-yl]-trimethyl-azanium | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with RT56
To Be Published
|
|
3ZRS
 
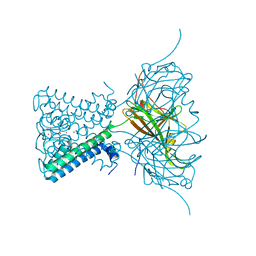 | | X-ray crystal structure of a KirBac potassium channel highlights a mechanism of channel opening at the bundle-crossing gate. | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION | | Authors: | Bavro, V.N, De Zorzi, R, Schmidt, M.R, Muniz, J.R.C, Zubcevic, L, Sansom, M.S.P, Venien-Bryan, C, Tucker, S.J. | | Deposit date: | 2011-06-17 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of a Kirbac Potassium Channel with an Open Bundle Crossing Indicates a Mechanism of Channel Gating
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ZCP
 
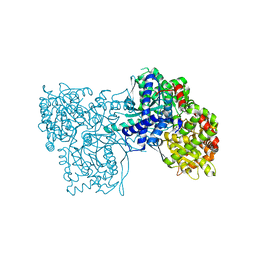 | | Rabbit muscle glycogen phosphorylase b in complex with N- cyclohexancarbonyl-N-beta-D-glucopyranosyl urea determined at 1.83 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(cyclohexylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZGF
 
 | | Crystal structure of the Fucosylgalactoside alpha N- acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with in complex with NPE caged UDP-Gal (P2(1)2(1)2(1) space group) | | Descriptor: | 1-(2-NITROPHENYL)ETHYL UDP-GALACTOSE, HISTO-BLOOD GROUP ABO SYSTEM TRANSFERASE, MANGANESE (II) ION, ... | | Authors: | Jorgensen, R, Batot, G.O, Hindsgaul, O, Tanaka, H, Perez, S, Imberty, A, Breton, C, Royant, A, Palcic, M.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structures of a Human Blood Group Glycosyltransferase in Complex with a Photo-Activatable Udp-Gal Derivative Reveal Two Different Binding Conformations
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7EPG
 
 | |
7EPI
 
 | |
7EPJ
 
 | |
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
6IBP
 
 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
3WYA
 
 | | Crystal structure of GDP-bound EF1alpha from Pyrococcus horikoshii | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
6ICG
 
 | | Grb2 SH2 domain in phosphopeptide free form | | Descriptor: | GLYCEROL, Growth factor receptor-bound protein 2, SULFATE ION | | Authors: | Hosoe, Y, Numoto, N, Inaba, S, Ogawa, S, Morii, H, Abe, R, Ito, N, Oda, M. | | Deposit date: | 2018-09-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and functional properties of Grb2 SH2 dimer in CD28 binding.
Biophys Physicobio., 16, 2019
|
|
3X1J
 
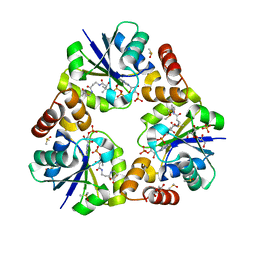 | |
1TYR
 
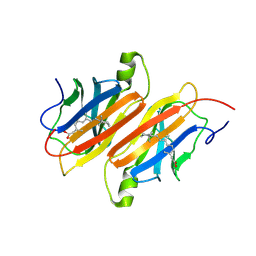 | | TRANSTHYRETIN COMPLEX WITH RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, TRANSTHYRETIN | | Authors: | Zanotti, G, D'Acunto, M.R, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1995-05-12 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the transthyretin--retinoic-acid complex
Eur.J.Biochem., 234, 1995
|
|
6I7O
 
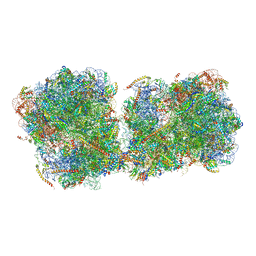 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2018-11-16 | | Release date: | 2019-01-16 | | Last modified: | 2019-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
6IOK
 
 | | Cryo-EM structure of multidrug efflux pump MexAB-OprM (0 degree state) | | Descriptor: | Multidrug resistance protein MexA, Multidrug resistance protein MexB, Outer membrane protein OprM | | Authors: | Tsutsumi, K, Yonehara, R, Nakagawa, A, Yamashita, E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2019-04-17 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of the wild-type MexAB-OprM tripartite pump reveal its complex formation and drug efflux mechanism.
Nat Commun, 10, 2019
|
|
3ZFF
 
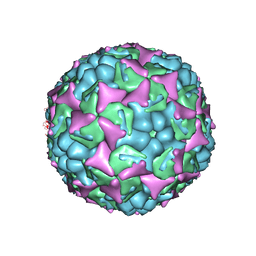 | | Human enterovirus 71 in complex with capsid binding inhibitor WIN51711 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, VP1, VP2, ... | | Authors: | Plevka, P, Perera, R, Yap, M.L, Cardosa, J, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2012-12-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Human Enterovirus 71 in Complex with a Capsid-Binding Inhibitor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
