5SNI
 
 | |
5SNE
 
 | |
5SNW
 
 | |
5SOA
 
 | |
5SNU
 
 | |
5SO8
 
 | |
5SN6
 
 | |
5SNN
 
 | |
5SO5
 
 | |
5SO1
 
 | |
5SOH
 
 | |
5SVF
 
 | | IDH1 R132H in complex with IDH125 | | Descriptor: | (4S)-3-(2-{[(1S)-1-phenylethyl]amino}pyrimidin-4-yl)-4-(propan-2-yl)-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
3H5Z
 
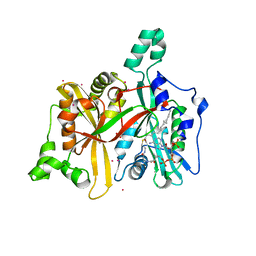 | | Crystal Structure of Leishmania major N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Qiu, W, Hutchinson, A, Lin, Y.-H, Wernimont, A, Mackenzie, F, Ravichandran, M, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Fairlamb, A.H, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | N-myristoyltransferase inhibitors as new leads to treat sleeping sickness.
Nature, 464, 2010
|
|
5XS8
 
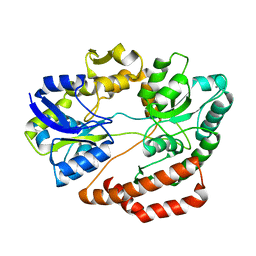 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Kamochi, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans.
Sci Rep, 7, 2017
|
|
4MZT
 
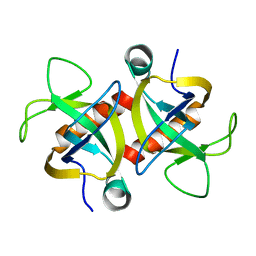 | | MazF from S. aureus crystal form II, C2221, 2.3 A | | Descriptor: | MazF mRNA interferase | | Authors: | Zorzini, V, Loris, R, van Nuland, N.A.J, Cheung, A. | | Deposit date: | 2013-09-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural and biophysical characterization of Staphylococcus aureus SaMazF shows conservation of functional dynamics.
Nucleic Acids Res., 42, 2014
|
|
5SZ2
 
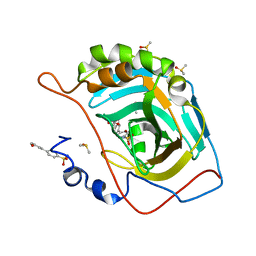 | | Carbonic anhydrase II in complex with 4-(3-formylphenyl)-benzenesulfonamide | | Descriptor: | 4-(3-formylphenyl)-benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Bhatt, A, Mahon, B.P, Cornelio, B, McKenna, R. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure-Activity Relationships of Benzenesulfonamide-Based Inhibitors towards Carbonic Anhydrase Isoform Specificity.
Chembiochem, 18, 2017
|
|
5SZ7
 
 | | Carbonic anhydrase IX-mimic in complex with 4-(3-quinolinyl)-benzenesulfonamide | | Descriptor: | 4-(3-quinolinyl)-benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Bhatt, A, Mahon, B.P, Cornelio, B, McKenna, R. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structure-Activity Relationships of Benzenesulfonamide-Based Inhibitors towards Carbonic Anhydrase Isoform Specificity.
Chembiochem, 18, 2017
|
|
5XQR
 
 | | Crystal structure of Notched-fin eelpout type III antifreeze protein A20V mutant (NFE6, AFP), C2221 form | | Descriptor: | ACETATE ION, Ice-structuring protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kondo, H, Tsuda, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Polypentagonal ice-like water networks emerge solely in an activity-improved variant of ice-binding protein
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5T15
 
 | | Structural basis for gating and activation of RyR1 (30 uM Ca2+ dataset, all particles) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1,Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6A2Q
 
 | | Mycobacterium tuberculosis LexA C-domain I | | Descriptor: | GLYCEROL, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5T4P
 
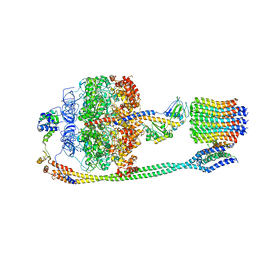 | | Autoinhibited E. coli ATP synthase state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.77 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
1IU5
 
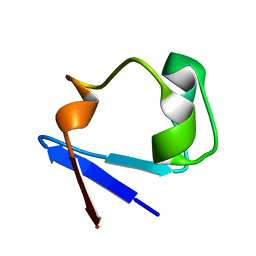 | | X-ray Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1IU6
 
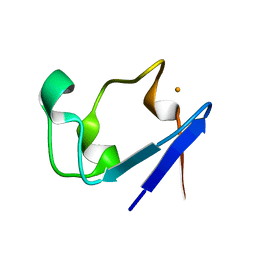 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3GW6
 
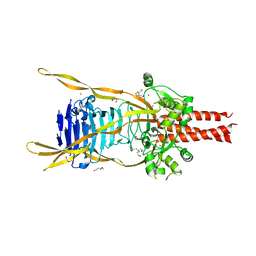 | | Intramolecular Chaperone | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schulz, E.C, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-03-31 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an intramolecular chaperone mediating triple-beta-helix folding.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4MV2
 
 | | Crystal structure of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Thomas, M.G, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-02 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
