5LGY
 
 | |
5LJF
 
 | | Crystal structure of the endo-1,4-glucanase RBcel1 E135A with cellotriose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutoit, R, Collet, L, Galleni, M, Bauvois, C. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.734396 Å) | | Cite: | Glycoside hydrolase family 5: structural snapshots highlighting the involvement of two conserved residues in catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5LFL
 
 | | MamA RS-1 ArsTM double mutant | | Descriptor: | CHLORIDE ION, Magnetosome protein MamA | | Authors: | Zarivach, R, Cronin, S.L. | | Deposit date: | 2016-07-03 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Differing self-assembling protein nanostructures in soluble and crystal form reveal dynamic behaviour
To Be Published
|
|
8HJZ
 
 | |
8HJX
 
 | |
8HJY
 
 | |
1OVB
 
 | | THE MECHANISM OF IRON UPTAKE BY TRANSFERRINS: THE STRUCTURE OF AN 18KD NII-DOMAIN FRAGMENT AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, OVOTRANSFERRIN | | Authors: | Kuser, P, Lindley, P, Sarra, R. | | Deposit date: | 1992-10-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of iron uptake by transferrins: the structure of an 18 kDa NII-domain fragment from duck ovotransferrin at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
8HUL
 
 | | X-ray structure of human PPAR delta ligand binding domain-lanifibranor co-crystals obtained by co-crystallization | | Descriptor: | 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUN
 
 | | X-ray structure of human PPAR alpha ligand binding domain-seladelpar co-crystals obtained by cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Iino, S, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUP
 
 | | X-ray structure of human PPAR gamma ligand binding domain-seladelpar-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, Isoform 1 of Peroxisome proliferator-activated receptor gamma, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUM
 
 | | X-ray structure of human PPAR gamma ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by co-crystallization | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Isoform 1 of Peroxisome proliferator-activated receptor gamma | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUO
 
 | | X-ray structure of human PPAR delta ligand binding domain-seladelpar co-crystals obtained by co-crystallization | | Descriptor: | Peroxisome proliferator-activated receptor delta, Seladelpar | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5LEV
 
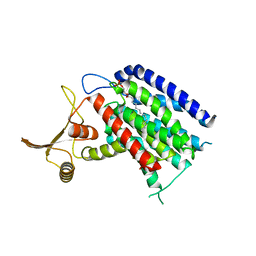 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | Descriptor: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
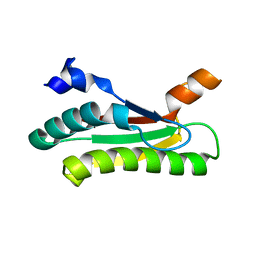
5LKW
 
 | |
5LTK
 
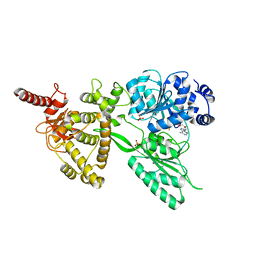 | |
5M0H
 
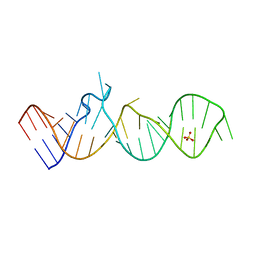 | |
3IWV
 
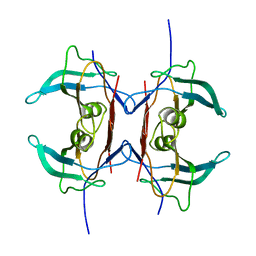 | | Crystal structure of Y116T mutant of 5-HYDROXYISOURATE HYDROLASE (TRP) | | Descriptor: | 5-hydroxyisourate hydrolase | | Authors: | Cendron, L, Ramazzina, I, Berni, R, Percudani, R, Zanotti, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Probing the evolution of hydroxyisourate hydrolase into transthyretin through active-site redesign.
J.Mol.Biol., 409, 2011
|
|
5LTJ
 
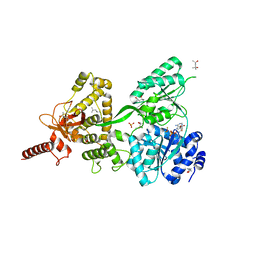 | | Crystal structure of the Prp43-ADP-BeF3 complex (in orthorhombic space group) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tauchert, M.J, Ficner, R. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the mechanism of the DEAH-box RNA helicase Prp43.
Elife, 6, 2017
|
|
8PJ0
 
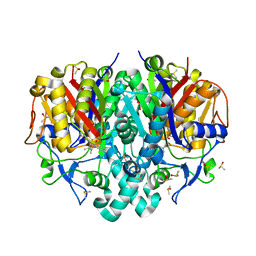 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-3-methylbutanamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
8PN5
 
 | |
8PN3
 
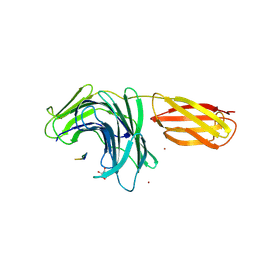 | |
5M0I
 
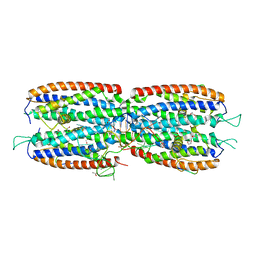 | | Crystal structure of the nuclear complex with She2p and the ASH1 mRNA E3-localization element | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASH1-E3 element, ... | | Authors: | Edelmann, F.T, Janowski, R, Niessing, D. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
8PMU
 
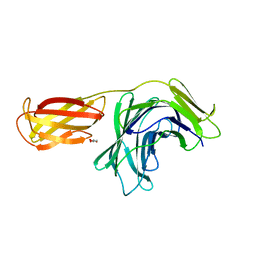 | |
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
