5QI0
 
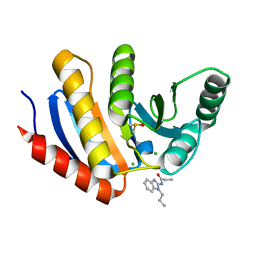 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QIW
 
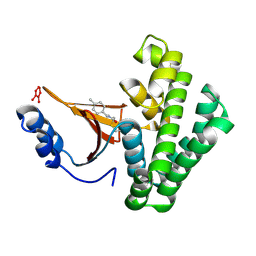 | | Covalent fragment group deposition -- Crystal Structure of OUTB2 in complex with PCM-0102660 | | Descriptor: | N-[(E)-(4-methylphenyl)methylidene]acetamide, UNKNOWN LIGAND, Ubiquitin thioesterase OTUB2 | | Authors: | Sethi, R, Douangamath, A, Resnick, E, Bradley, A.R, Collins, P, Brandao-Neto, J, Talon, R, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, London, N, von Delft, F. | | Deposit date: | 2018-08-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalent fragment group deposition
To Be Published
|
|
2Z2J
 
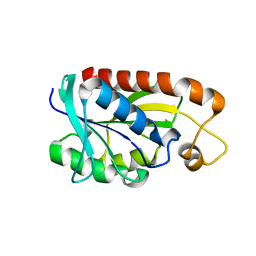 | | Crystal structure of Peptidyl-tRNA hydrolase from Mycobacterium tuberculosis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Selvaraj, M, Roy, S, Singh, N.S, Sangeetha, R, Varshney, U, Vijayan, M. | | Deposit date: | 2007-05-22 | | Release date: | 2007-07-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Plasticity and Enzyme Action: Crystal Structures of Mycobacterium tuberculosis Peptidyl-tRNA Hydrolase
J.Mol.Biol., 372, 2007
|
|
2Z44
 
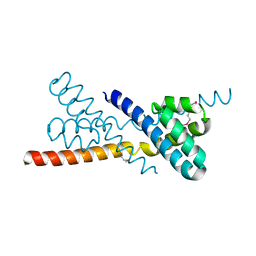 | | Crystal Structure of Selenomethionine-labeled ORF134 | | Descriptor: | ORF134 | | Authors: | Tomimoto, Y, Ihara, K, Onizuka, T, Kanai, S, Ashida, H, Yokota, A, Tanaka, S, Miyasaka, H, Yamada, Y, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of ORF134
To be Published
|
|
4C10
 
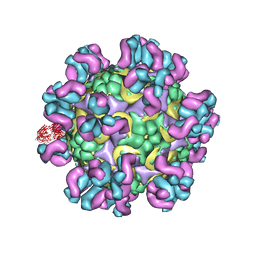 | | Cryo-EM reconstruction of empty enterovirus 71 in complex with a neutralizing antibody E19 | | Descriptor: | CHLORIDE ION, EV19 5 C1-6 F1 C11, SODIUM ION, ... | | Authors: | Plevka, P, Perera, R, Cardosa, J, Suksatu, A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2013-08-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2Z87
 
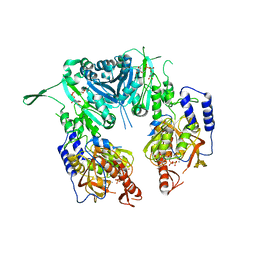 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
19HC
 
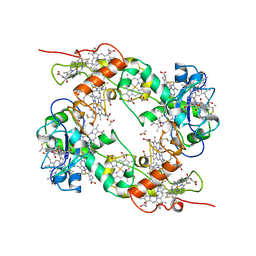 | | NINE-HAEM CYTOCHROME C FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ACETATE ION, PROTEIN (NINE-HAEM CYTOCHROME C), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Coelho, R, Pereira, I.A.C, Coelho, A.V, Thompson, A.W, Sieker, L, Gall, J.L, Carrondo, M.A. | | Deposit date: | 1998-12-01 | | Release date: | 1999-12-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family.
Structure Fold.Des., 7, 1999
|
|
2Z52
 
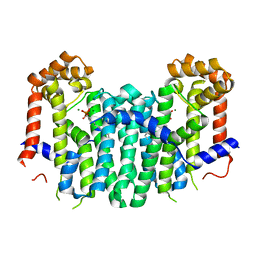 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-23 | | Descriptor: | (1-HYDROXYDODECANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Chen, C.K.-M, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
117E
 
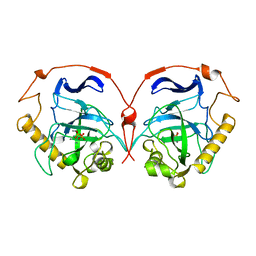 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-15 | | Release date: | 1998-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
2ZAM
 
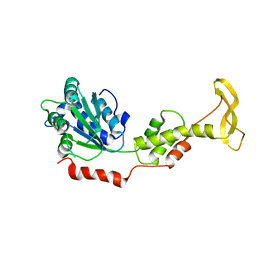 | | Crystal structure of mouse SKD1/VPS4B apo-form | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Inoue, M, Kawasaki, M, Kamikubo, H, Kataoka, M, Kato, R, Yoshimori, T, Wakatsuki, S. | | Deposit date: | 2007-10-08 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B
Traffic, 9, 2008
|
|
4GBD
 
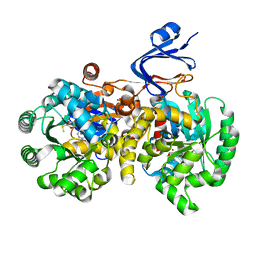 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
5QHW
 
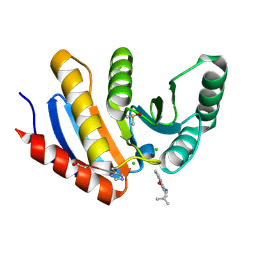 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000347a | | Descriptor: | 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1J6Y
 
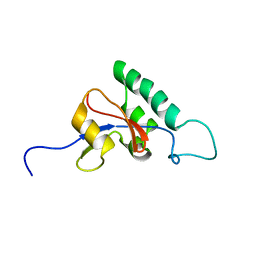 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
4IXU
 
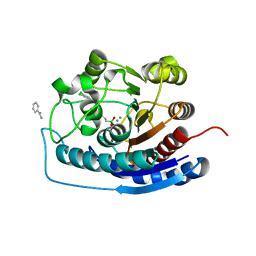 | | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1J80
 
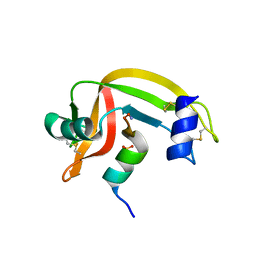 | | Osmolyte Stabilization of RNase | | Descriptor: | RIBONUCLEASE PANCREATIC, SULFATE ION | | Authors: | Ratnaparkhi, G.S, Varadarajan, R. | | Deposit date: | 2001-05-19 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Osmolytes stabilize ribonuclease S by stabilizing its fragments S protein and S peptide to compact folding-competent states.
J.Biol.Chem., 276, 2001
|
|
2Z4X
 
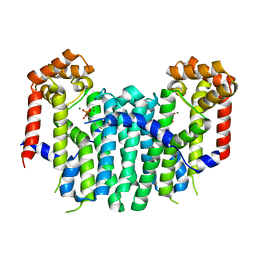 | | S. cerevisiae geranylgeranyl pyrophosphate synthase in complex with magnesium and BPH-252 (P21) | | Descriptor: | (1-HYDROXYNONANE-1,1-DIYL)BIS(PHOSPHONIC ACID), Geranylgeranyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Guo, R.T, Chen, C.K.-M, Hudock, M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-06-26 | | Release date: | 2008-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of geranylgeranyl diphosphate synthase by bisphosphonates: a crystallographic and computational investigation
J.Med.Chem., 51, 2008
|
|
133D
 
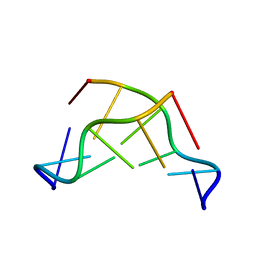 | | THE CRYSTAL STRUCTURE OF N4-METHYLCYTOSINE.GUANOSIN BASE-PAIRS IN THE SYNTHETIC HEXANUCLEOTIDE D(CGCGM(4)CG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C34)P*G)-3') | | Authors: | Cervi, A.R, Guy, A, Leonard, G.A, Teoule, R, Hunter, W.N. | | Deposit date: | 1993-07-29 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of N4-methylcytosine.guanosine base-pairs in the synthetic hexanucleotide d(CGCGm4CG).
Nucleic Acids Res., 21, 1993
|
|
2HEI
 
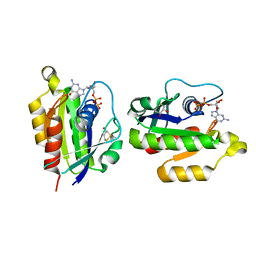 | | Crystal structure of human RAB5B in complex with GDP | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-5B | | Authors: | Hong, B, Shen, L, Wang, J, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of human RAB5B in complex with GDP
To be Published
|
|
1II5
 
 | |
4BZJ
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4C0U
 
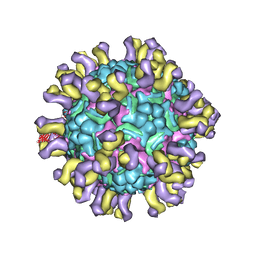 | | Cryo-EM reconstruction of enterovirus 71 in complex with a neutralizing antibody E18 | | Descriptor: | FAB EV18 4 D6-1 F1 G9, VP1, VP2, ... | | Authors: | Plevka, P, Perera, R, Cardosa, J, Suksatu, A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2013-08-08 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Neutralizing Antibodies Can Initiate Genome Release from Human Enterovirus 71.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1IK0
 
 | |
5QHJ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human FAM83B in complex with FMOPL000709a | | Descriptor: | (2S)-1-{[(2H-1,3-benzodioxol-5-yl)methyl]amino}propan-2-ol, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Talon, R, Krojer, T, Douangamath, A, Collins, P, Zhang, R, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bullock, A.N. | | Deposit date: | 2018-05-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2Z67
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) selenium transferase (SepSecS) | | Descriptor: | O-phosphoseryl-tRNA(Sec) selenium transferase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Araiso, Y, Ishitani, R, Pailouer, S, Oshikane, H, Domae, N, Soll, D, Nureki, O. | | Deposit date: | 2007-07-23 | | Release date: | 2008-04-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation.
Nucleic Acids Res., 36, 2008
|
|
4BN9
 
 | |
