4BAC
 
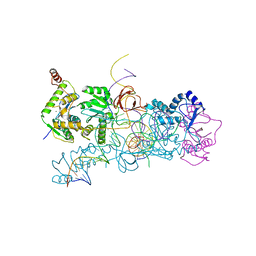 | | prototype foamy virus strand transfer complexes on product DNA | | Descriptor: | 5'-D(*AP*GP*GP*AP*GP*CP*CP*AP*AP*GP*AP*CP*GP*GP *AP*TP*CP)-3', 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*TP*GP*TP*AP)-3', DNA (38-MER), ... | | Authors: | Yin, Z, Lapkouski, M, Yang, W, Craigie, R. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Protein Sci., 21, 2012
|
|
4BDD
 
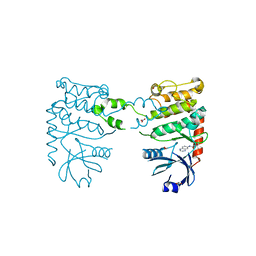 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 1-(TERT-BUTYL)-3-(QUINOXALIN-6-YL)UREA, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4AXS
 
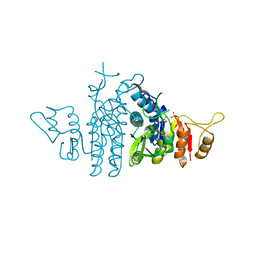 | | Structure of Carbamate Kinase from Mycoplasma penetrans | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Gallego, P, Planell, R, Benach, J, Querol, E, PerezPons, J.A, Reverter, D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4AY7
 
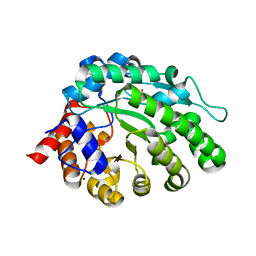 | | methyltransferase from Methanosarcina mazei | | Descriptor: | MAGNESIUM ION, METHYLCOBALAMIN: COENZYME M METHYLTRANSFERASE, ZINC ION | | Authors: | Hoeppner, A, Thomas, F, Rueppel, A, Hensel, R, Blankenfeld, W, Bayer, P, Faust, A. | | Deposit date: | 2012-06-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Corrinoid:Coenzyme M Methyltransferase Mtaa from Methanosarcina Mazei
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B0G
 
 | | Complex of Aurora-A bound to an Imidazopyridine-based inhibitor | | Descriptor: | 6-bromo-2-(1-methyl-1H-imidazol-5-yl)-7-{4-[(5-methyl-1,2-oxazol-3-yl)methyl]piperazin-1-yl}-1H-imidazo[4,5-b]pyridine, AURORA KINASE A, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2012-07-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Optimization of Imidazo[4,5-B]Pyridine-Based Kinase Inhibitors: Identification of a Dual Flt3/Aurora Kinase Inhibitor as an Orally Bioavailable Preclinical Development Candidate for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 55, 2012
|
|
4BG4
 
 | | Crystal structure of Litopenaeus vannamei arginine kinase in a ternary analog complex with arginine, ADP-Mg and NO3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, ARGININE KINASE, ... | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, GBrieba, L, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-03-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
6JKM
 
 | | Crystal structure of BubR1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
4B9E
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | Descriptor: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | Authors: | Schmidberger, J.W, Schnell, R, Schneider, G. | | Deposit date: | 2012-09-04 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4BEB
 
 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
4B3U
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-amino-1-benzyl-5-(ethylamino)pyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
4B4D
 
 | | Crystal structure of FAD-containing ferredoxin-NADP reductase from Xanthomonas axonopodis pv. citri | | Descriptor: | CHLORIDE ION, FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Martinez-Julvez, M, Orellano, E.G, Tondo, M.L, Hurtado-Guerrero, R, Medina, M, Ceccarelli, E.A, Sanchez-Azqueta, A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Fad-Containing Ferredoxin- Nadp Reductase from Xanthomonas Axonopodis Pv. Citri
Biomed Res Int, 2013, 2013
|
|
4B5C
 
 | | Crystal structure of the peptidoglycan-associated lipoprotein from Burkholderia pseudomallei | | Descriptor: | ACETATE ION, PUTATIVE OMPA FAMILY LIPOPROTEIN | | Authors: | Gourlay, L.J, Peri, C, Conchillo-Sole, O, Ferrer-Navarro, M, Gori, A, Longhi, R, Rinchai, D, Lertmemongkolchai, G, Lassaux, P, Daura, X, Colombo, G, Bolognesi, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Burkholderia Pseudomallei Acute Phase Antigen Bpsl2765 for Structure-Based Epitope Discovery/Design in Structural Vaccinology.
Chem.Biool., 20, 2013
|
|
6JEC
 
 | |
3ZFO
 
 | | Crystal structure of substrate-like, unprocessed N-terminal protease Npro mutant S169P | | Descriptor: | CHLORIDE ION, HYDROXIDE ION, MONOTHIOGLYCEROL, ... | | Authors: | Zogg, T, Sponring, M, Schindler, S, Koll, M, Schneider, R, Brandstetter, H, Auer, B. | | Deposit date: | 2012-12-12 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structures of the Viral Protease Npro Imply Distinct Roles for the Catalytic Water in Catalysis
Structure, 21, 2013
|
|
6JKL
 
 | |
6JKQ
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi (Ligand-free form) | | Descriptor: | Aspartate carbamoyltransferase | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
3ZN1
 
 | | LSD1-CoREST in complex with PRLYLV peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, PEPTIDE, ... | | Authors: | Tortorici, M, Borrello, M.T, Tardugno, M, Chiarelli, L.R, Pilotto, S, Ciossani, G, Vellore, N.A, Cowan, J, O'Connell, M, Mai, A, Baron, R, Ganesan, A, Mattevi, A. | | Deposit date: | 2013-02-13 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Recognition by Small Peptide Reversible Inhibitors of the Chromatin-Modifying Lsd1/Corest Lysine Demethylase.
Acs Chem.Biol., 8, 2013
|
|
3ZCD
 
 | |
6JL6
 
 | | Crystal structure of aspartate transcarbamoylase from Trypanosoma cruzi in complex with phosphate (Pi). | | Descriptor: | ASPARTIC ACID, Aspartate carbamoyltransferase, PHOSPHATE ION | | Authors: | Matoba, K, Shiba, T, Nara, T, Aoki, T, Nagasaki, S, Hayamizu, R, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Balogun, E.O, Inaoka, D.K, Kita, K, Harada, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshots of Trypanosoma cruzi aspartate transcarbamoylase
revealed an ordered Bi-Bi reaction mechanism
To Be Published
|
|
6JOO
 
 | | Crystal structure of Corynebacterium diphtheriae Cas9 in complex with sgRNA and target DNA | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated protein,CRISPR-associated endonuclease Cas9, Guide RNA, ... | | Authors: | Hirano, S, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2019-03-22 | | Release date: | 2019-04-17 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the promiscuous PAM recognition by Corynebacterium diphtheriae Cas9.
Nat Commun, 10, 2019
|
|
1UWP
 
 | |
3ZL5
 
 | | Crystal structure of Schistosoma mansoni Peroxiredoxin I C48S mutant with one decamer in the ASU | | Descriptor: | DI(HYDROXYETHYL)ETHER, PEROXIREDOXIN I, SULFATE ION | | Authors: | Saccoccia, F, Angelucci, F, Ardini, M, Boumis, G, Brunori, M, DiLeandro, L, Ippoliti, R, Miele, A.E, Natoli, G, Scotti, S, Bellelli, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Switching between the Alternative Structures and Functions of a 2-Cys Peroxiredoxin, by Site-Directed Mutagenesis
J.Mol.Biol., 425, 2013
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
6K0P
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
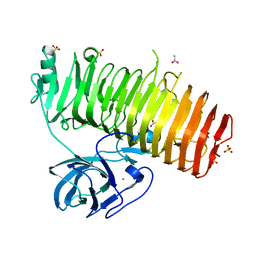 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
