1DD3
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
6LVB
 
 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3T0O
 
 | | Crystal Structure Analysis of Human RNase T2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribonuclease T2 | | Authors: | Thorn, A, Kraetzner, R, Steinfeld, R, Sheldrick, G. | | Deposit date: | 2011-07-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and activity of the only human RNase T2.
Nucleic Acids Res., 40, 2012
|
|
4LOB
 
 | | Crystal structure of polyprenyl diphosphate synthase A1S_2732 (Target EFI-509223) from Acinetobacter baumannii | | Descriptor: | CHLORIDE ION, GLYCEROL, Polyprenyl synthetase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of polyprenyl diphosphate synthase from Acinetobacter baumannii
To be Published
|
|
6ZLY
 
 | |
1DJ3
 
 | | STRUCTURES OF ADENYLOSUCCINATE SYNTHETASE FROM TRITICUM AESTIVUM AND ARABIDOPSIS THALIANA | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Prade, L, Cowan-Jacob, S.W, Chemla, P, Potter, S, Ward, E, Fonne-Pfister, R. | | Deposit date: | 1999-12-01 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of adenylosuccinate synthetase from Triticum aestivum and Arabidopsis thaliana.
J.Mol.Biol., 296, 2000
|
|
1DM1
 
 | | 2.0 A CRYSTAL STRUCTURE OF THE DOUBLE MUTANT H(E7)V, T(E10)R OF MYOGLOBIN FROM APLYSIA LIMACINA | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Federici, L, Savino, C, Musto, R, Travaglini-Allocatelli, C, Cutruzzola, F, Brunori, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering His(E7) affects the control of heme reactivity in Aplysia limacina myoglobin.
Biochem.Biophys.Res.Commun., 269, 2000
|
|
1DMK
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
3KLV
 
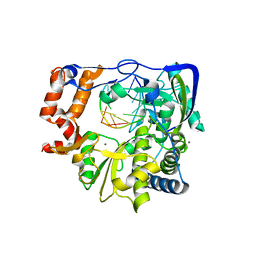 | | M296I G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA | | Descriptor: | 3D polymerase, MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*GP*GP*CP*C)-3'), ... | | Authors: | Ferrer-Orta, C, Sierra, M, Agudo, R, Perez-Luque, R, Arias, A, Verdaguer, N. | | Deposit date: | 2009-11-09 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
1DQ6
 
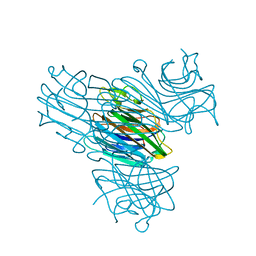 | | Manganese;Manganese concanavalin A at pH 7.0 | | Descriptor: | Concanavalin-A, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
5UAE
 
 | | Crystal structure of the coiled-coil domain from Listeria Innocua Phage Integrase (Trigonal Form) | | Descriptor: | CITRATE ANION, Putative integrase | | Authors: | Gupta, K, Sharp, R, Yuan, J.B, Van Duyne, G.D. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coiled-coil interactions mediate serine integrase directionality.
Nucleic Acids Res., 45, 2017
|
|
1QRB
 
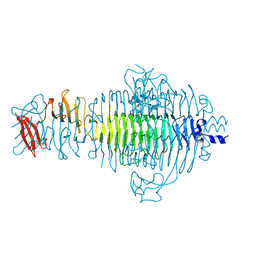 | | PLASTICITY AND STERIC STRAIN IN A PARALLEL BETA-HELIX: RATIONAL MUTATIONS IN P22 TAILSPIKE PROTEIN | | Descriptor: | PROTEIN (TAILSPIKE-PROTEIN) | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-12 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
5UDO
 
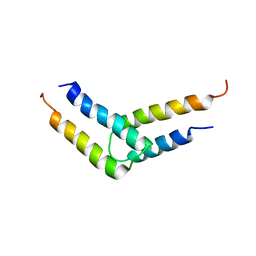 | |
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
1E4K
 
 | | CRYSTAL STRUCTURE OF SOLUBLE HUMAN IGG1 FC FRAGMENT-FC-GAMMA RECEPTOR III COMPLEX | | Descriptor: | FC FRAGMENT OF HUMAN IGG1, LOW AFFINITY IMMUNOGLOBULIN GAMMA FC RECEPTOR III, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sondermann, P, Huber, R, Oosthuizen, V, Jacob, U. | | Deposit date: | 2000-07-07 | | Release date: | 2000-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The 3.2A Crystal Structure of the Human Igg1 Fc Fragment-Fc-Gamma-Riii Complex
Nature, 406, 2000
|
|
3JT0
 
 | | Crystal Structure of the C-terminal fragment (426-558) Lamin-B1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR5546A | | Descriptor: | Lamin-B1 | | Authors: | Kuzin, A, Abashidze, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-09-22 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR5546A
To be Published
|
|
3ZY5
 
 | | Crystal structure of POFUT1 in complex with GDP-fucose (crystal-form-I) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, PUTATIVE GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 1, ... | | Authors: | Lira-Navarrete, E, Valero-Gonzalez, J, Villanueva, R, Martinez-Julvez, M, Tejero, T, Merino, P, Panjikar, S, Hurtado-Guerrero, R. | | Deposit date: | 2011-08-17 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Insights Into the Mechanism of Protein O-Fucosylation.
Plos One, 6, 2011
|
|
1DMG
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L4 | | Descriptor: | CITRIC ACID, RIBOSOMAL PROTEIN L4 | | Authors: | Worbs, M, Huber, R, Wahl, M.C. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ribosomal protein L4 shows RNA-binding sites for ribosome incorporation and feedback control of the S10 operon.
EMBO J., 19, 2000
|
|
1DO3
 
 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T>180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
4EEK
 
 | | Crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) With bound phosphate and sodium | | Descriptor: | Beta-phosphoglucomutase-related protein, PHOSPHATE ION, SODIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
4OO7
 
 | | THE 1.55A CRYSTAL STRUCTURE of NAF1 (MINER1): THE REDOX-ACTIVE 2FE-2S PROTEIN | | Descriptor: | CDGSH iron-sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Tamir, S, Eisenberg-Domovich, Y, Colman, A.R, Stofleth, J.T, Lipper, C.H, Paddock, M.L, Jenning, P.A, Livnah, O, Nechushtai, R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A point mutation in the [2Fe-2S] cluster binding region of the NAF-1 protein (H114C) dramatically hinders the cluster donor properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3OWQ
 
 | | X-Ray Structure of Lin1025 protein from Listeria innocua, Northeast Structural Genomics Consortium Target LkR164 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1025 protein | | Authors: | Kuzin, A, Su, M, Lew, S, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-20 | | Release date: | 2010-10-20 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR164
To be published
|
|
4OOK
 
 | | Third Metal bound M.tuberculosis methionine aminopeptidase | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 2, SODIUM ION | | Authors: | Reddi, R, Addlagatta, A. | | Deposit date: | 2014-02-03 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective targeting of the conserved active site cysteine of Mycobacterium tuberculosis methionine aminopeptidase with electrophilic reagents
Febs J., 281, 2014
|
|
1DPF
 
 | | CRYSTAL STRUCTURE OF A MG-FREE FORM OF RHOA COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RHOA | | Authors: | Shimizu, T, Ihara, K, Maesaki, R, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1999-12-27 | | Release date: | 2000-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An open conformation of switch I revealed by the crystal structure of a Mg2+-free form of RHOA complexed with GDP. Implications for the GDP/GTP exchange mechanism.
J.Biol.Chem., 275, 2000
|
|
1DQW
 
 | | CRYSTAL STRUCTURE OF OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Descriptor: | OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Milburn, M.V, Miller, B.G, Hassell, A.M, Wolfenden, R, Short, S.A. | | Deposit date: | 2000-01-05 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anatomy of a proficient enzyme: the structure of orotidine 5'-monophosphate decarboxylase in the presence and absence of a potential transition state analog.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
