7AV8
 
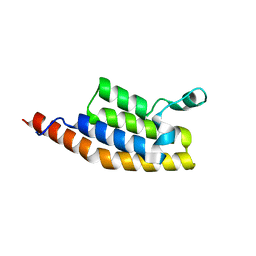 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7B36
 
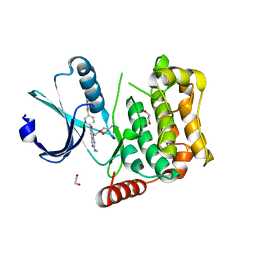 | | MST4 in complex with compound G-5555 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, ... | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.10681081 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2WVQ
 
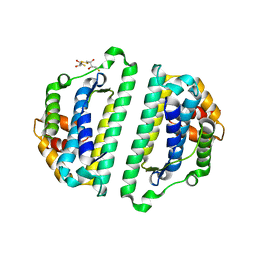 | | Structure of the HET-s N-terminal domain. Mutant D23A, P33H | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SMALL S PROTEIN | | Authors: | Greenwald, J, Buhtz, C, Ritter, C, Kwiatkowski, W, Choe, S, Saupe, S.J, Riek, R. | | Deposit date: | 2009-10-19 | | Release date: | 2010-07-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of prion inhibition by HET-S.
Mol. Cell, 38, 2010
|
|
7VTI
 
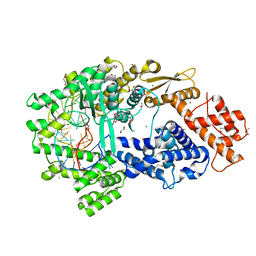 | | Crystal structure of the Cas13bt3-crRNA binary complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
7B6I
 
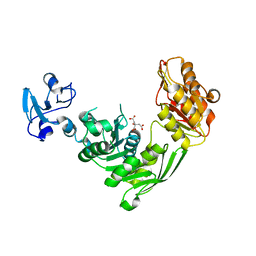 | | Crystal structure of MurE from E.coli in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
2X58
 
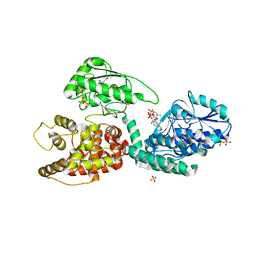 | | The crystal structure of MFE1 liganded with CoA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, GLYCEROL, ... | | Authors: | Kasaragod, P, Venkatesan, R, Kiema, T.R, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Liganded Rat Peroxisomal Multifunctional Enzyme Type 1: A Flexible Molecule with Two Interconnected Active Sites
J.Biol.Chem., 285, 2010
|
|
7B7N
 
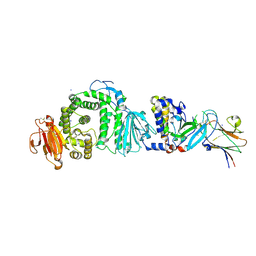 | | Human herpesvirus-8 gH/gL in complex with EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pederzoli, R, Guardado-Calvo, P, Rey, F.A, Backovic, M. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Human herpesvirus 8 molecular mimicry of ephrin ligands facilitates cell entry and triggers EphA2 signaling.
Plos Biol., 19, 2021
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7B5E
 
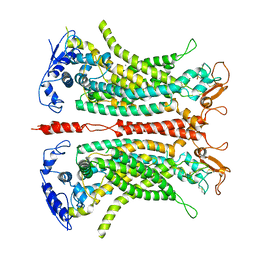 | | Structure of calcium-bound mTMEM16A(ac)-I551A chloride channel at 4.1 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7B5D
 
 | |
2XB0
 
 | |
7VQF
 
 | | Phenol binding protein, MopR | | Descriptor: | ACETATE ION, PHENOL, Phenol sensing regulator, ... | | Authors: | Singh, J, Ray, S, Anand, R. | | Deposit date: | 2021-10-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenol sensing in nature is modulated via a conformational switch governed by dynamic allostery.
J.Biol.Chem., 298, 2022
|
|
7B5C
 
 | | Structure of calcium-bound mTMEM16A(ac) chloride channel at 3.7 A resolution | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Lam, A.K.M, Rheinberger, J, Paulino, C, Dutzler, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-02-10 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Gating the pore of the calcium-activated chloride channel TMEM16A.
Nat Commun, 12, 2021
|
|
7AJR
 
 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
2X1T
 
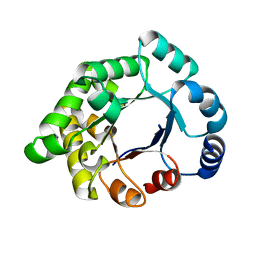 | | Crystallographic binding studies with an engineered monomeric variant of triosephosphate isomerase | | Descriptor: | 4-PHOSPHO-D-ERYTHRONOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Salin, M, Kapetaniou, E.G, Vaismaa, M, Lajunen, M, Casteleijn, M.G, Neubauer, P, Salmon, L, Wierenga, R. | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic Binding Studies with an Engineered Monomeric Variant of Triosephosphate Isomerase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2WRA
 
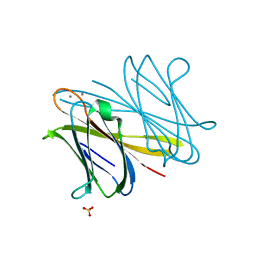 | | BclA lectin from Burkholderia cenocepacia complexed with aMan1(aMan1- 6)-3Man trisaccharide | | Descriptor: | CALCIUM ION, LECTIN, SULFATE ION, ... | | Authors: | Lameignere, E, Shiao, T.C, Roy, R, Wimmerova, M, Dubreuil, F, Varrot, A, Imberty, A. | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Basis of the Affinity for Oligomannosides and Analogs Displayed by Bc2L-A, a Burkholderia Cenocepacia Soluble Lectin.
Glycobiology, 20, 2010
|
|
2WU4
 
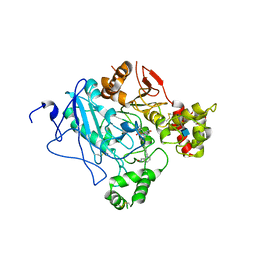 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND ORTHO-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
2WZF
 
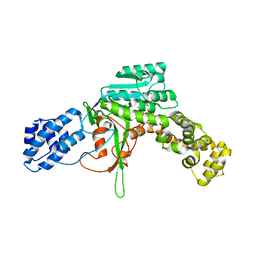 | | Legionella pneumophila glucosyltransferase crystal structure | | Descriptor: | GLUCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Hurtado-Guerrero, R, Zusman, T, Pathak, S, Ibrahim, A.F.M, Shepherd, S, Prescott, A, Segal, G, van Aalten, D.M.F. | | Deposit date: | 2009-11-29 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Elongation Factor 1A Inhibition by a Legionella Pneumophila Glycosyltransferase.
Biochem.J., 426, 2010
|
|
7VTN
 
 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
2X4F
 
 | | The Crystal Structure of the human myosin light chain kinase LOC340156. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-(3-dioxaziridin-3-ylphenyl)pyrimidin-2-amine, MYOSIN LIGHT CHAIN KINASE FAMILY MEMBER 4, ... | | Authors: | Muniz, J.R.C, Mahajan, P, Rellos, P, Fedorov, O, Shrestha, B, Wang, J, Elkins, J.M, Daga, N, Cocking, R, Chaikuad, A, Krojer, T, Ugochukwu, E, Yue, W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Gileadi, O, Knapp, S. | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | The Crystal Structure of the Human Myosin Light Chain Kinase Loc340156
To be Published
|
|
2X7N
 
 | | Mechanism of eIF6s anti-association activity | | Descriptor: | 60S RIBOSOMAL PROTEIN L23, 60S RIBOSOMAL PROTEIN L24-A, EUKARYOTIC TRANSLATION INITIATION FACTOR 6, ... | | Authors: | Gartmann, M, Blau, M, Armache, J.-P, Mielke, T, Topf, M, Beckmann, R. | | Deposit date: | 2010-03-02 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | Mechanism of Eif6-Mediated Inhibition of Ribosomal Subunit Joining.
J.Biol.Chem., 285, 2010
|
|
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | Descriptor: | ACETYL COENZYME *A, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
7VB1
 
 | | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-08-30 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | The 0.90 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with trans-vaccenic acid
To Be Published
|
|
2WSQ
 
 | | MonoTIM mutant RMM0-1, dimeric form. | | Descriptor: | SULFATE ION, TRIOSE PHOSPHATE ISOMERASE, GLYCOSOMAL | | Authors: | Rudino-Pinera, E, Rojas-Trejo, S.P, Arreola, R, Saab-Rincon, G, Soberon, X, Horjales, E. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Space Group Transition Driven by Temperature and Related to Monomer-Dimer Transition in Solution: The Case of Monomeric Tim of Trypanosoma Brucei Brucei
To be Published
|
|
7B51
 
 | | Crystal structure of human CRM1 covalently modified by 2-mercaptoethanol at Cys528 | | Descriptor: | BETA-MERCAPTOETHANOL, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-12-03 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of human CRM1, covalently modified by 2-mercaptoethanol on Cys528, in complex with RanGTP.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
