5IMH
 
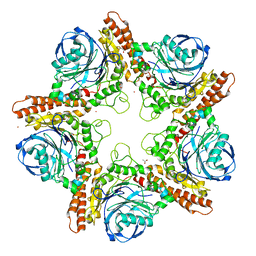 | |
5IN3
 
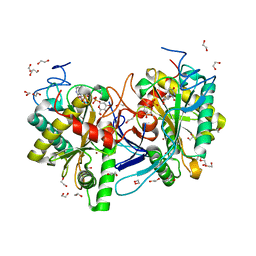 | | Crystal structure of glucose-1-phosphate bound nucleotidylated human galactose-1-phosphate uridylyltransferase | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Kopec, J, McCorvie, T, Tallant, C, Velupillai, S, Shrestha, L, Fitzpatrick, F, Patel, D, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular basis of classic galactosemia from the structure of human galactose 1-phosphate uridylyltransferase.
Hum.Mol.Genet., 25, 2016
|
|
5INH
 
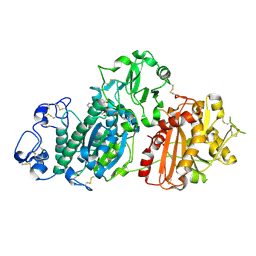 | | Crystal structure of Autotaxin/ENPP2 with a covalent fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Klein, M.G, Tjhen, R. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Repurposing Suzuki Coupling Reagents as a Directed Fragment Library Targeting Serine Hydrolases and Related Enzymes.
J. Med. Chem., 60, 2017
|
|
5IQO
 
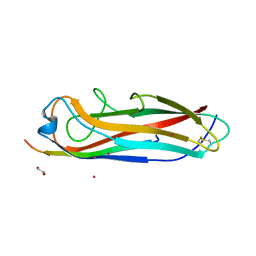 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitutions Q134E and S138E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Giese, C, Eras, J, Kern, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5IQW
 
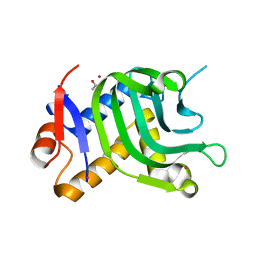 | | 1.95A resolution structure of Apo HasAp (R33A) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, CADMIUM ION, Heme acquisition protein HasAp | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Yao, H, Rivera, M. | | Deposit date: | 2016-03-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Replacing Arginine 33 for Alanine in the Hemophore HasA from Pseudomonas aeruginosa Causes Closure of the H32 Loop in the Apo-Protein.
Biochemistry, 55, 2016
|
|
5IMQ
 
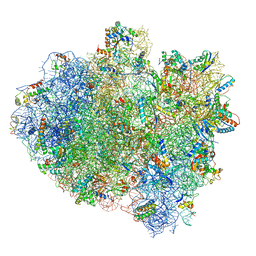 | | Structure of ribosome bound to cofactor at 3.8 angstrom resolution | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Kumar, V, Ero, R, Jian, G.K, Ahmed, T, Zhan, Y, Bhushan, S, Gao, Y.G. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the GTP Form of Elongation Factor 4 (EF4) Bound to the Ribosome
J.Biol.Chem., 291, 2016
|
|
5IQX
 
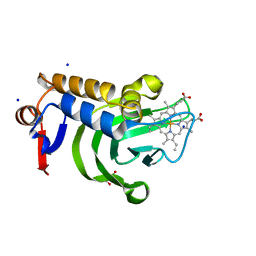 | | 1.05A resolution structure of Holo HasAp (R33A) from Pseudomonas aeruginosa | | Descriptor: | D-MALATE, Heme acquisition protein HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kumar, R, Lovell, S, Battaile, K.P, Yao, H, Rivera, M. | | Deposit date: | 2016-03-11 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Replacing Arginine 33 for Alanine in the Hemophore HasA from Pseudomonas aeruginosa Causes Closure of the H32 Loop in the Apo-Protein.
Biochemistry, 55, 2016
|
|
5IRA
 
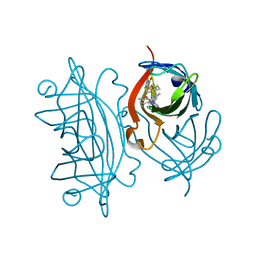 | | Expanding Nature's Catalytic Repertoire -Directed Evolution of an Artificial Metalloenzyme for In Vivo Metathesis | | Descriptor: | Artificial Metathesase, [1-[4-[[5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]methyl]-2,6-dimethyl-phenyl]-3-(2,4,6-trimethylphenyl)-4,5-dihydroimidazol-1-ium-2-yl]-bis(chloranyl)ruthenium | | Authors: | Heinisch, T, Jeschek, M, Reuter, R, Trindler, C, Panke, S, Ward, T.R. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Directed evolution of artificial metalloenzymes for in vivo metathesis.
Nature, 537, 2016
|
|
3QXO
 
 | | CDK2 in complex with inhibitor KVR-1-84 | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-2-[(4-sulfamoylbenzyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-02 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QRT
 
 | | CDK2 in complex with inhibitor NSK-MC2-55 | | Descriptor: | (5R)-5-(2-methylbutan-2-yl)-N-(4-sulfamoylbenzyl)-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
5I5A
 
 | | quasi racemic structure of allo-Ile7-ShK and D-ShK | | Descriptor: | D-MALATE, D-ShK, GLYCEROL, ... | | Authors: | Dang, B, Kubota, T, Mandal, K, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2JCN
 
 | | The crystal structure of BAK1 - a mitochondrial apoptosis regulator | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, SULFATE ION | | Authors: | Moche, M, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Lundgren, S, Nilsson, M.E, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-27 | | Release date: | 2007-01-04 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Bak1 - an Apoptosis Trigger in the Mitochondrial Outer Membrane
To be Published
|
|
5I69
 
 | | MBP-MamC magnetite-interaction component mutant-D70A | | Descriptor: | Maltose-binding periplasmic protein,Tightly bound bacterial magnetic particle protein,Maltose-binding periplasmic protein, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nudelman, H, Tercedor, C.V, Kolusheva, S, Gonzalez, T.P, Widdrat, M, Grimberg, N, Levi, H, Nelkenbaum, O, Davidove, G, Faivre, D, Jimenez-Lopez, C, Zarivach, R. | | Deposit date: | 2016-02-16 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC.
J.Struct.Biol., 194, 2016
|
|
5ZXF
 
 | | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Cheng, X.Y, Su, Z.D, Pi, N, Cao, C.Z, Zhao, Z.T, Zhou, J.J, Chen, R, Kuang, Z.K, Huang, Y.Q. | | Deposit date: | 2018-05-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a
To Be Published
|
|
2ZE4
 
 | | Crystal structure of phospholipase D from streptomyces antibioticus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Phospholipase D | | Authors: | Suzuki, A, Kakuno, K, Saito, R, Iwasaki, Y, Yamane, T, Yamane, T. | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of phospholipase D from streptomyces antibioticus
To be Published
|
|
4LP0
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
3QQL
 
 | | CDK2 in complex with inhibitor L3 | | Descriptor: | (5R)-5-(2-methylbutan-2-yl)-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
5INR
 
 | |
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
3QWJ
 
 | | CDK2 in complex with inhibitor KVR-1-142 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-chloro-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-28 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QX4
 
 | | CDK2 in complex with inhibitor KVR-1-78 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
5I6D
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 5 [3-(3-(p-Tolyl)ureido) benzoic acid] | | Descriptor: | 3-{[(4-methylphenyl)carbamoyl]amino}benzoic acid, GLYCEROL, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-16 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
5I7A
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 1 [3-(3-(3,4-Dichlorophenyl)ureido)benzoic acid] | | Descriptor: | 3-{[(3,4-dichlorophenyl)carbamoyl]amino}benzoic acid, O-phosphoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-17 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
5I94
 
 | | Crystal structure of human glutaminase C in complex with the inhibitor UPGL-00019 | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2016-02-19 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
3QX2
 
 | | CDK2 in complex with inhibitor KVR-1-190 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-4-[(2-hydroxyethyl)amino]-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
