5CBP
 
 | | Crystal Structure of Conjoint Pyrococcus furiosus L-asparaginase at 37 degree C | | Descriptor: | CITRATE ANION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Sharma, P, Yadav, S.P, Tomar, R, Kundu, B, Ashish, F. | | Deposit date: | 2015-07-01 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Heat induces end to end repetitive association in P. furiosus L-asparaginase which enables its thermophilic property.
Sci Rep, 10, 2020
|
|
2MWQ
 
 | | Solution structure of PsbQ from spinacia oleracea | | Descriptor: | Oxygen-evolving enhancer protein 3, chloroplastic | | Authors: | Rathner, P, Mueller, N, Wimmer, R, Chandra, K. | | Deposit date: | 2014-11-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants.
Proteins, 83, 2015
|
|
5CKF
 
 | | E. coli MazF E24A form I | | Descriptor: | Endoribonuclease MazF, SODIUM ION | | Authors: | Zorzini, V, Loris, R. | | Deposit date: | 2015-07-15 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate Recognition and Activity Regulation of the Escherichia coli mRNA Endonuclease MazF.
J.Biol.Chem., 291, 2016
|
|
4QOT
 
 | | Crystal structure of human copper chaperone bound to the platinum ion | | Descriptor: | Copper transport protein ATOX1, PLATINUM (II) ION, SULFATE ION | | Authors: | Belviso, B.D, Galliani, A, Caliandro, R, Arnesano, F, Natile, G. | | Deposit date: | 2014-06-20 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxaliplatin Binding to Human Copper Chaperone Atox1 and Protein Dimerization
Inorg.Chem., 55, 2016
|
|
4QK3
 
 | |
4QPB
 
 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | Descriptor: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
7KI0
 
 | | Semaglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs protein | | Descriptor: | 17-amino-10-oxo-3,6,12,15-tetraoxa-9-azaheptadecan-1-oic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
2I14
 
 | |
5M5W
 
 | | RNA Polymerase I open complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-07 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
7KI1
 
 | | Taspoglutide-bound Glucagon-Like Peptide-1 (GLP-1) Receptor in Complex with Gs Protein | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, X, Belousoff, M.J, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2020-10-22 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and dynamics of semaglutide- and taspoglutide-bound GLP-1R-Gs complexes.
Cell Rep, 36, 2021
|
|
3R6X
 
 | | CDK2 in complex with inhibitor KVR-1-158 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-hydroxypropyl)amino]-5-nitro-2-[(pyridin-3-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-22 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
3R7U
 
 | | CDK2 in complex with inhibitor KVR-1-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(aminomethyl)benzyl]amino}-5-nitrobenzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
4QME
 
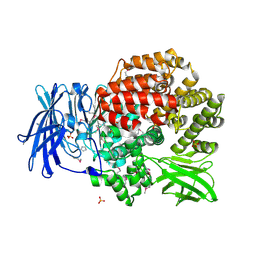 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe | | Descriptor: | (2S)-3-[(S)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Vassilious, S, Mulligan, R, Berlicki, L, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QNS
 
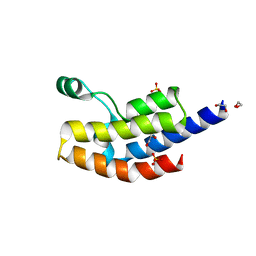 | | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase GCN5, ... | | Authors: | Fonseca, M, Tallant, C, Knapp, S, Loppnau, P, von Delft, F, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Crystal structure of bromodomain from Plasmodium faciparum GCN5, PF3D7_0823300
TO BE PUBLISHED
|
|
4QNY
 
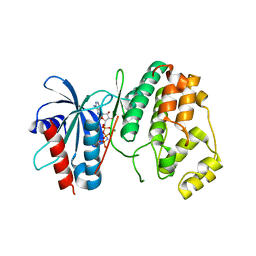 | | Crystal structure of MapK from Leishmania donovani, LDBPK_331470 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mitogen activated protein kinase, ... | | Authors: | Wernimont, A.K, Loppnau, P, Walker, J.R, Mangos, M, El Bakkouri, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Crystal structure of MapK from Leishmania donovani, LDBPK_331470
TO BE PUBLISHED
|
|
5MEG
 
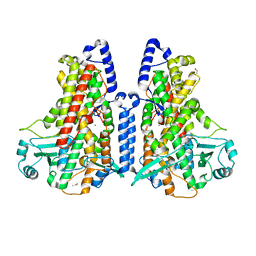 | | Manganese-substituted Cyanothece lipoxygenase 2 (Mn-CspLOX2) | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, ETHANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
4QVS
 
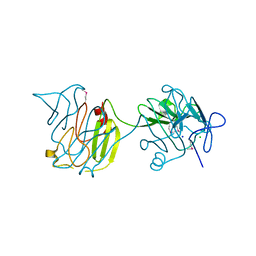 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
5MEY
 
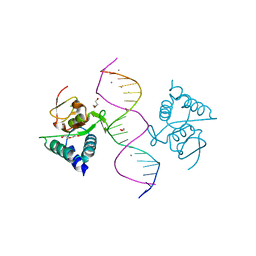 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
3RPO
 
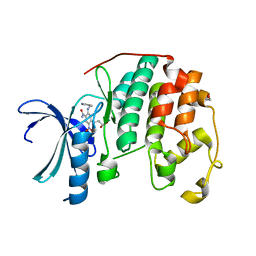 | | CDK2 in complex with inhibitor KVR-1-156 | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-carbamoyl-2-nitro-5-[(pyridin-3-ylmethyl)amino]phenyl}amino)butanoic acid, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-27 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
5C65
 
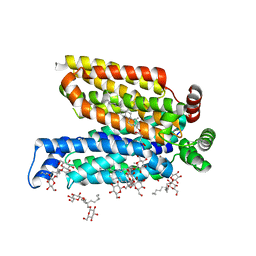 | | Structure of the human glucose transporter GLUT3 / SLC2A3 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Octyl Glucose Neopentyl Glycol, Solute carrier family 2, ... | | Authors: | Pike, A.C.W, Quigley, A, Chu, A, Tessitore, A, Xia, X, Mukhopadhyay, S, Wang, D, Kupinska, K, Strain-Damerell, C, Chalk, R, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-22 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the human glucose transporter GLUT3 / SLC2A3
To Be Published
|
|
5C6L
 
 | | Crystal Structure of Gadolinium derivative of HEWL solved using intense Free-Electron Laser radiation | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | Galli, L, Barends, T.R.M, Son, S.-K, White, T.A, Barty, A, Botha, S, Boutet, S, Caleman, C, Doak, R.B, Nanao, M.H, Nass, K, Shoeman, R.L, Timneanu, N, Santra, R, Schlichting, I, Chapman, H.N. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards phasing using high X-ray intensity.
Iucrj, 2, 2015
|
|
4QYM
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, MAGNESIUM ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with methionine
To be Published
|
|
5M5Y
 
 | | RNA Polymerase I elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Hoffmann, N.A, Jakobi, A.J, Wetzel, R, Hagen, W.J.H, Sachse, C, Muller, C.W. | | Deposit date: | 2016-10-23 | | Release date: | 2016-12-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular Structures of Transcribing RNA Polymerase I.
Mol. Cell, 64, 2016
|
|
3R7V
 
 | | CDK2 in complex with inhibitor KVR-1-9 | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-2-[(pyridin-2-ylmethyl)amino]benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-23 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
