4ZHO
 
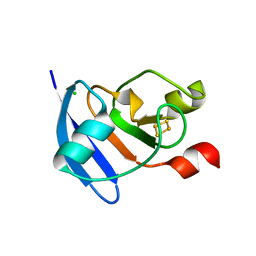 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
5L07
 
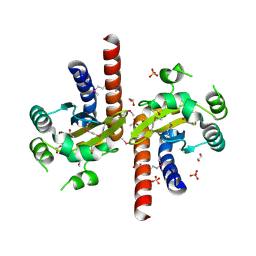 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Quorum-sensing transcriptional activator, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KPO
 
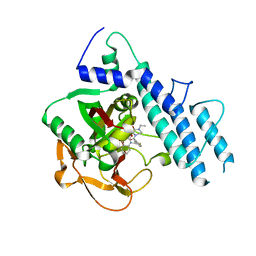 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
1HUQ
 
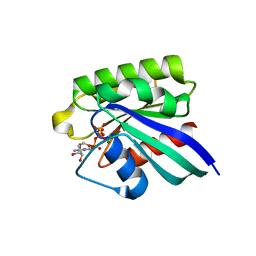 | | 1.8A CRYSTAL STRUCTURE OF THE MONOMERIC GTPASE RAB5C (MOUSE) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB5C | | Authors: | Merithew, E, Hatherly, S, Dumas, J.J, Lawe, D.C, Heller-Harrison, R, Lambright, D.G. | | Deposit date: | 2001-01-04 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition.
J.Biol.Chem., 276, 2001
|
|
3V04
 
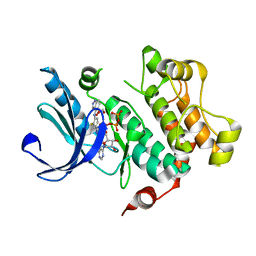 | | Discovery of Novel Allosteric MEK Inhibitors Possessing Classical and Non-classical Bidentate Ser212 Interactions. | | Descriptor: | 4-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-1H-indazole-5-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Heald, R, Jackson, P, Savy, P, Jones, M, Gancia, E, Burton, B, Newman, R, Boggs, J, Chan, E, Chan, J, Choo, E, Merchant, M, Ultsch, M, Wiesmann, C, Belvin, M, Price, S. | | Deposit date: | 2011-12-07 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Novel Allosteric Mitogen-Activated Protein Kinase Kinase (MEK) 1,2 Inhibitors Possessing Bidentate Ser212 Interactions.
J.Med.Chem., 55, 2012
|
|
2K75
 
 | | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum. Northeast Structural Genomics Consortium target TaR80b. | | Descriptor: | uncharacterized protein Ta0387 | | Authors: | Ramelot, T.A, Ding, K, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-01 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the OB domain of Ta0387 from Thermoplasma acidophilum.
To be Published
|
|
1HNR
 
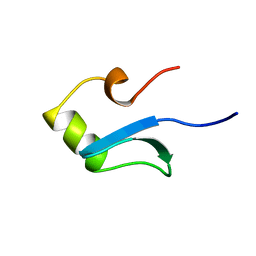 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
4ZO9
 
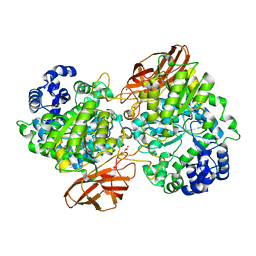 | | Crystal Structure of mutant (D270A) beta-glucosidase from Listeria innocua in complex with laminaribiose | | Descriptor: | GLYCEROL, Lin1840 protein, MAGNESIUM ION, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
5KJX
 
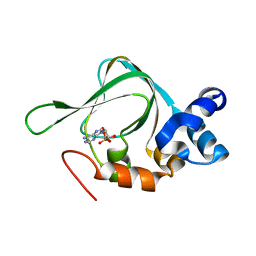 | | Co-crystal Structure of PKA RI alpha CNB-B domain with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Lorenz, R, Moon, E, Kim, J.J, Huang, G.Y, Kim, C, Herberg, F.W. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutations of PKA cyclic nucleotide-binding domains reveal novel aspects of cyclic nucleotide selectivity.
Biochem. J., 474, 2017
|
|
1HJN
 
 | | HUMAN PRION PROTEIN AT PH 7.0 | | Descriptor: | MAJOR PRION PROTEIN PRECURSOR | | Authors: | Calzolai, L, Zahn, R. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Influence of Ph on NMR Structure and Stability of the Human Prion Protein Globular Domain
J.Biol.Chem., 278, 2003
|
|
1HO7
 
 | | NMR STRUCTURE OF THE POTASSIUM CHANNEL FRAGMENT L45 IN TFE | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN | | Authors: | Ohlenschlager, O, Hojo, H, Ramachandran, R, Gorlach, M, Haris, P.I. | | Deposit date: | 2000-12-10 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the S4-S5 segment of the Shaker potassium channel.
Biophys.J., 82, 2002
|
|
2KCQ
 
 | | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855) . Northeast Structural Genomics Consortium target SrR106 | | Descriptor: | Mov34/MPN/PAD-1 family | | Authors: | Wu, Y, Eletsky, A, Zhao, L, Hua, J, Sukumaran, D, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-28 | | Release date: | 2009-02-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein SRU_2040 from Salinibacter ruber (strain DSM 13855). Northeast Structural Genomics Consortium target SrR106
To be Published
|
|
4YY8
 
 | | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum | | Descriptor: | GLYCEROL, Kelch protein, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, He, H, Ravichandran, M, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, El Bakkouri, M, Senisterra, G, Osman, K.T, Lovato, D.V, Hui, R, Hutchinson, A, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure Analysis of Kelch protein from Plasmodium falciparum.
to be published
|
|
2KCK
 
 | | NMR solution structure of the Northeast Structural Genomics Consortium (NESG) target MrR121A | | Descriptor: | TPR repeat | | Authors: | Barb, A.W, Lee, H.-W, Wang, X, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Northeast Structural Genomics Target MrR121A
To be Published
|
|
1HLE
 
 | | CRYSTAL STRUCTURE OF CLEAVED EQUINE LEUCOCYTE ELASTASE INHIBITOR DETERMINED AT 1.95 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, HORSE LEUKOCYTE ELASTASE INHIBITOR | | Authors: | Baumann, U, Bode, W, Huber, R, Travis, J, Potempa, J. | | Deposit date: | 1992-04-13 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of cleaved equine leucocyte elastase inhibitor determined at 1.95 A resolution.
J.Mol.Biol., 226, 1992
|
|
5I7R
 
 | | Mycobacterium tuberculosis CysM in complex with the Urea-scaffold inhibitor 2 [3-(3-([1,1'-biphenyl]-3-yl)ureido)benzoic acid] | | Descriptor: | 3-{[([1,1'-biphenyl]-3-yl)carbamoyl]amino}benzoic acid, ACETATE ION, O-phosphoserine sulfhydrylase, ... | | Authors: | Schnell, R, Maric, S, Schneider, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis.
J.Med.Chem., 59, 2016
|
|
4Z36
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2015-07-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
5I47
 
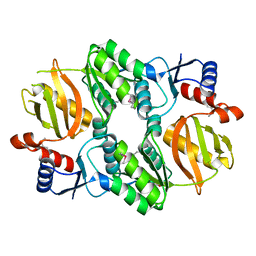 | | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, RimK domain protein ATP-grasp | | Authors: | Chang, C, Duke, N, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745
To Be Published
|
|
5IAU
 
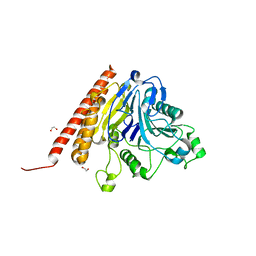 | |
4ZNV
 
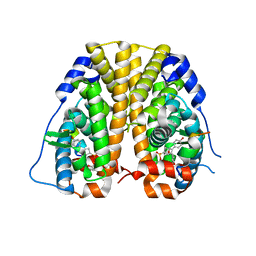 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with a 2-Methoxy-substituted OBHS derivative | | Descriptor: | 2-methoxyphenyl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate, Estrogen receptor, Nuclear receptor-interacting peptide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-05 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4ZOA
 
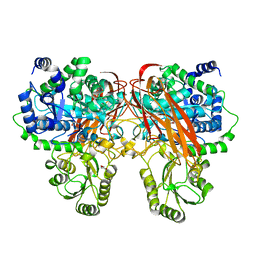 | | Crystal Structure of beta-glucosidase from Listeria innocua in complex with isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, DI(HYDROXYETHYL)ETHER, Lin1840 protein, ... | | Authors: | Nakajima, M, Yoshida, R, Miyanaga, A, Abe, K, Takahashi, Y, Sugimoto, N, Toyoizumi, H, Nakai, H, Kitaoka, M, Taguchi, H. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Functional and Structural Analysis of a beta-Glucosidase Involved in beta-1,2-Glucan Metabolism in Listeria innocua
Plos One, 11, 2016
|
|
5ICG
 
 | | Crystal structure of apo (S)-norcoclaurine 6-O-methyltransferase | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, POTASSIUM ION | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
5I5C
 
 | | X-ray crystal structure of allo-Thr31-ShK | | Descriptor: | GLYCEROL, Kappa-stichotoxin-She3a, LITHIUM ION, ... | | Authors: | Dang, B, Kubota, T, Manda, K.l, Shen, R, Bezanilla, F, Roux, B, Kent, S.B.H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Inversion of the Side-Chain Stereochemistry of Indvidual Thr or Ile Residues in a Protein Molecule: Impact on the Folding, Stability, and Structure of the ShK Toxin.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2KGR
 
 | | Solution structure of protein ITSN1 from Homo sapiens. Northeast Structural Genomics Consortium target HR5524A | | Descriptor: | Intersectin-1 | | Authors: | Wu, Y, Ghosh, A, Shastry, R, Hua, J, Ciccosanti, C, Zhang, Q, Jiang, M, Swapna, G, Acton, T, Xiao, R, Everett, J, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-16 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein ITSN1 from Homo sapiens. Northeast Structural Genomics Consortium target HR5524A
To be Published
|
|
4ZPO
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Rubinstein, R, Honig, B, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
