3WS4
 
 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WU8
 
 | |
3WU7
 
 | |
1CQA
 
 | | BIRCH POLLEN PROFILIN | | Descriptor: | PROFILIN | | Authors: | Fedorov, A.A, Ball, T, Mahoney, N.M, Valenta, R, Almo, S.C. | | Deposit date: | 1996-07-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin.
Structure, 5, 1997
|
|
3NRW
 
 | | Crystal Structure of the N-terminal domain of Phage integrase/site-specific recombinase (tnp) from Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR208A | | Descriptor: | Phage integrase/site-specific recombinase | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Northeast Structural Genomics Consortium Target HmR208A
To be Published
|
|
4EXJ
 
 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
1D4U
 
 | | INTERACTIONS OF HUMAN NUCLEOTIDE EXCISION REPAIR PROTEIN XPA WITH RPA70 AND DNA: CHEMICAL SHIFT MAPPING AND 15N NMR RELAXATION STUDIES | | Descriptor: | NUCLEOTIDE EXCISION REPAIR PROTEIN XPA (XPA-MBD), ZINC ION | | Authors: | Buchko, G.W, Daughdrill, G.W, de Lorimier, R, Rao, S, Isern, N.G, Lingbeck, J, Taylor, J, Wold, M.S, Gochin, M, Spicer, L.D, Lowry, D.F, Kennedy, M.A. | | Deposit date: | 1999-10-06 | | Release date: | 1999-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies.
Biochemistry, 38, 1999
|
|
1D5A
 
 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK. DEPOSITION OF SECOND NATIVE STRUCTURE AT 2.4 ANGSTROM | | Descriptor: | MAGNESIUM ION, PROTEIN (DNA POLYMERASE), SULFATE ION | | Authors: | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | Deposit date: | 1999-10-06 | | Release date: | 2000-03-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaebacterial DNA polymerase.
Structure Fold.Des., 7, 1999
|
|
1CQW
 
 | | NAI COCRYSTALLISED WITH HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE; 1-CHLOROHEXANE HALIDOHYDROLASE, IODIDE ION | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1999-08-11 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1D75
 
 | | CONFORMATION OF THE GUANINE.8-OXOADENINE BASE PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(O8A)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(A38)P*GP*CP*G)-3') | | Authors: | Leonard, G.A, Guy, A, Brown, T, Teoule, R, Hunter, W.N. | | Deposit date: | 1992-05-07 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation of guanine-8-oxoadenine base pairs in the crystal structure of d(CGCGAATT(O8A)GCG).
Biochemistry, 31, 1992
|
|
3O3W
 
 | | Crystal Structure of BH2092 protein (residues 14-131) from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228A | | Descriptor: | BH2092 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-26 | | Release date: | 2010-09-01 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal Structure of BH2092 protein (residues 14-131) from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR228A
To be Published
|
|
6KXM
 
 | |
4HWU
 
 | | Crystal structure of the Ig-C2 type 1 domain from mouse Fibroblast growth factor receptor 2 (FGFR2) [NYSGRC-005912] | | Descriptor: | Fibroblast growth factor receptor 2 | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of the Ig-C2 type 1 domain from mouse FGFR2 [NYSGRC-005912]
to be published
|
|
3WRT
 
 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WUA
 
 | |
3WUH
 
 | | Qri7 and AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, tRNA N6-adenosine threonylcarbamoyltransferase, ... | | Authors: | Tominaga, T, Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
4NAV
 
 | | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608 | | Descriptor: | HYPOTHETICAL PROTEIN XCC279 | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Zhao, S.C, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Stead, M, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of hypothetical protein XCC2798 from Xanthomonas campestris, Target EFI-508608
TO BE PUBLISHED
|
|
3KB2
 
 | | Crystal Structure of YorR protein in complex with phosphorylated GDP from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR256 | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION, SPBc2 prophage-derived uncharacterized protein yorR | | Authors: | Forouhar, F, Friedman, D, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Ho, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR256
To be Published
|
|
1D66
 
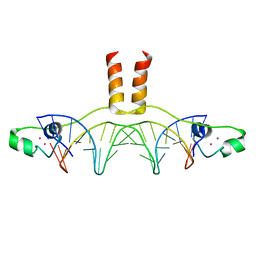 | | DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*AP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*AP*GP*GP*AP*CP*TP*GP*TP*CP*CP*TP*CP*C P*GP*G)-3'), ... | | Authors: | Marmorstein, R, Carey, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1992-03-06 | | Release date: | 1992-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | DNA recognition by GAL4: structure of a protein-DNA complex.
Nature, 356, 1992
|
|
1CG9
 
 | | COMPLEX RECOGNITION OF THE SUPERTYPIC BW6-DETERMINANT ON HLA-B AND-C MOLECULES BY THE MONOCLONAL ANTIBODY SFR8-B6 | | Descriptor: | PROTEIN (BETA-2-MICROGLOBULIN), PROTEIN (EBNA-6 NUCLEAR PROTEIN (EBNA-3C) (EBNA-4B)), PROTEIN (HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Menssen, R, Orth, P, Ziegler, A, Saenger, W. | | Deposit date: | 1999-03-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Recognition of the Supertypic Bw6-Determinant on HLA-B and-C Molecules
by the Monoclonal Antibody SFR8-B6
To be Published
|
|
3KAV
 
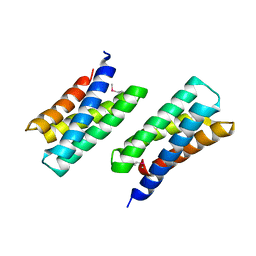 | | Crystal Structure of THE MUTANT (L80M) PA2107 PROTEIN from Pseudomonas aeruginosa, Northeast Structural Genomics Consortium Target PaR198 | | Descriptor: | uncharacterized protein PA2107 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target PaR198
To be Published
|
|
1D1D
 
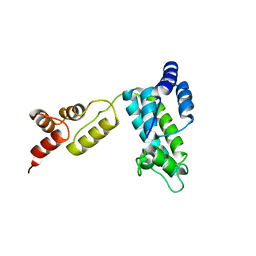 | |
1NM7
 
 | | Solution structure of the ScPex13p SH3 domain | | Descriptor: | Peroxisomal Membrane Protein PAS20 | | Authors: | Pires, J.R, Hong, X, Brockmann, C, Volkmer-Engert, R, Schneider-Mergener, J, Oschkinat, H, Erdmann, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The ScPex13p SH3 Domain Exposes Two Distinct Binding Sites for Pex5p and Pex14p
J.Mol.Biol., 326, 2003
|
|
3W90
 
 | |
1CTT
 
 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE NUCLEOSIDE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
