2FMN
 
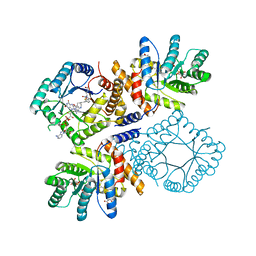 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase complex with LY309887 | | 分子名称: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, N-[(5-{2-[(6R)-2-AMINO-4-OXO-3,4,5,6,7,8-HEXAHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL]ETHYL}-2-THIENYL)CARBONYL]-L-GLUTAMIC ACID | | 著者 | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | 登録日 | 2006-01-09 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
4UWD
 
 | |
2FPN
 
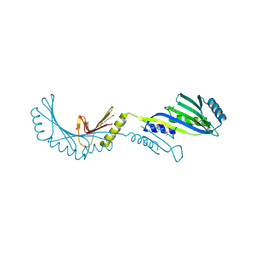 | |
9JDW
 
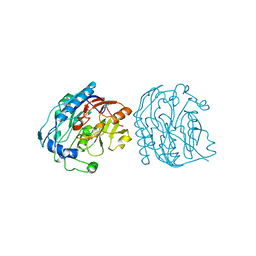 | |
2FMO
 
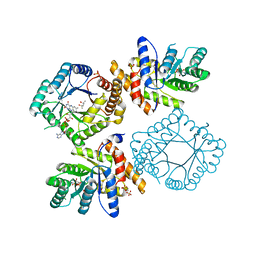 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | 登録日 | 2006-01-09 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
2FQ4
 
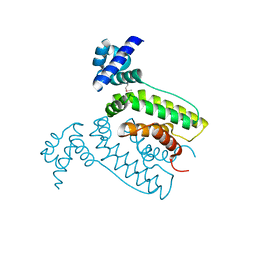 | | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus | | 分子名称: | Transcriptional regulator, TetR family | | 著者 | Zhang, R, Wu, R, Moy, S, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-01-17 | | 公開日 | 2006-02-28 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | The crystal structure of the transcriptional regulator (TetR family) from Bacillus cereus
To be Published
|
|
2FQ3
 
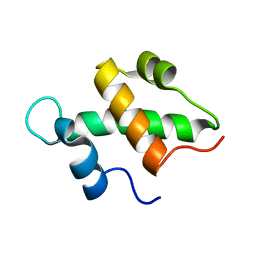 | | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | | 分子名称: | Transcription regulatory protein SWI3 | | 著者 | Da, G, Lenkart, J, Zhao, K, Shiekhattar, R, Cairns, B.R, Marmorstein, R. | | 登録日 | 2006-01-17 | | 公開日 | 2006-02-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4V6I
 
 | | Localization of the small subunit ribosomal proteins into a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein RACK1 (RACK1), ... | | 著者 | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | 登録日 | 2010-10-12 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.8 Å) | | 主引用文献 | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4UY9
 
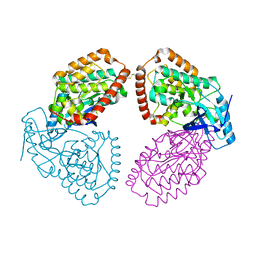 | | Structure of MLK1 kinase domain with leucine zipper 1 | | 分子名称: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 9 | | 著者 | Read, J.A, Brassington, C, Pollard, H.K, Phillips, C, Green, I, Overmann, R, Collier, M. | | 登録日 | 2014-08-29 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Recurrent Mlk4 Loss-of-Function Mutations Suppress Jnk Signaling to Promote Colon Tumorigenesis.
Cancer Res., 76, 2016
|
|
3H3Q
 
 | |
5VE4
 
 | | Crystal structure of persulfide dioxygenase-rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans | | 分子名称: | BpPRF, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | 登録日 | 2017-04-03 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | 分子名称: | Phospholipase A2 VRV-PL-VIIIa | | 著者 | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | 登録日 | 2017-04-05 | | 公開日 | 2018-06-20 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VIS
 
 | | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil Uncultured Bacterium. | | 分子名称: | CHLORIDE ION, D(-)-TARTARIC ACID, Dihydropteroate Synthase, ... | | 著者 | Minasov, G, Wawrzak, Z, Di Leo, R, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-04-17 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | 1.73 Angstrom Resolution Crystal Structure of Dihydropteroate Synthase (folP-SMZ_B27) from Soil
Uncultured Bacterium.
To Be Published
|
|
6Q3S
 
 | | Engineered Human HLA_A2 MHC Class I molecule in complex with TCR and SV9 peptide | | 分子名称: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Meijers, R, Anjanappa, R, Springer, S, Garcia-Alai, M. | | 登録日 | 2018-12-04 | | 公開日 | 2019-07-24 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | High-throughput peptide-MHC complex generation and kinetic screenings of TCRs with peptide-receptive HLA-A*02:01 molecules.
Sci Immunol, 4, 2019
|
|
5VE5
 
 | | Crystal structure of persulfide dioxygenase rhodanese fusion protein with rhodanese domain inactivating mutation (C314S) from Burkholderia phytofirmans in complex with glutathione | | 分子名称: | BpPRF, CHLORIDE ION, FE (III) ION, ... | | 著者 | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | 登録日 | 2017-04-03 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
3GVK
 
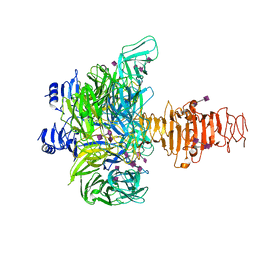 | | Crystal structure of endo-neuraminidase NF mutant | | 分子名称: | Endo-N-acetylneuraminidase, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid, ... | | 著者 | Schulz, E.C, Dickmanns, A, Ficner, R. | | 登録日 | 2009-03-31 | | 公開日 | 2010-03-02 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural basis for the recognition and cleavage of polysialic acid by the bacteriophage K1F tailspike protein EndoNF.
J.Mol.Biol., 397, 2010
|
|
3H3R
 
 | |
5VTG
 
 | |
5VYJ
 
 | |
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | 登録日 | 2017-04-05 | | 公開日 | 2017-11-29 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VIP
 
 | |
5VF5
 
 | | Crystal structure of the vicilin from Solanum melongena, re-refinement | | 分子名称: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Porebski, P.J, Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | 登録日 | 2017-04-06 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VIT
 
 | |
5VE3
 
 | | Crystal structure of wild-type persulfide dioxygenase-rhodanese fusion protein from Burkholderia phytofirmans | | 分子名称: | BpPRF, FE (III) ION | | 著者 | Motl, N, Skiba, M.A, Smith, J.L, Banerjee, R. | | 登録日 | 2017-04-03 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.793 Å) | | 主引用文献 | Structural and biochemical analyses indicate that a bacterial persulfide dioxygenase-rhodanese fusion protein functions in sulfur assimilation.
J. Biol. Chem., 292, 2017
|
|
5VSV
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | 著者 | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-05-12 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.205 Å) | | 主引用文献 | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
