7NBU
 
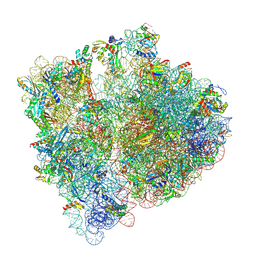 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | 登録日 | 2021-01-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | 登録日 | 2020-02-15 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
5XEZ
 
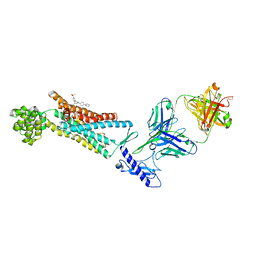 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | 著者 | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2017-04-06 | | 公開日 | 2017-05-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
1N6D
 
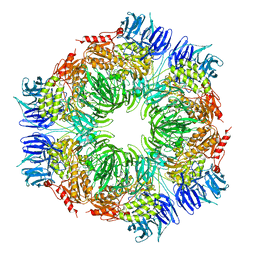 | | Tricorn protease in complex with tetrapeptide chloromethyl ketone derivative | | 分子名称: | DECANE, RVRK, Tricorn protease | | 著者 | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | 登録日 | 2002-11-10 | | 公開日 | 2002-12-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
6Y3C
 
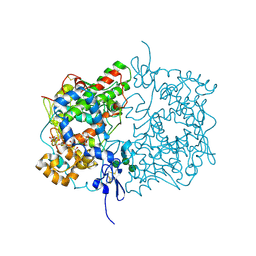 | | Human COX-1 Crystal Structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Prostaglandin G/H synthase 1, ... | | 著者 | Miciaccia, M, Belviso, B.D, Iaselli, M, Ferorelli, S, Perrone, M.G, Caliandro, R, Scilimati, A. | | 登録日 | 2020-02-18 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.361 Å) | | 主引用文献 | Three-dimensional structure of human cyclooxygenase (hCOX)-1.
Sci Rep, 11, 2021
|
|
5XFU
 
 | | Domain swapped dimer crystal structure of loop1 deletion mutant in Single-chain Monellin | | 分子名称: | Monellin chain B,Monellin chain A | | 著者 | Surana, P, Nandwani, N, Udgaonkar, J, Gosavi, S, Das, R. | | 登録日 | 2017-04-11 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.611 Å) | | 主引用文献 | Amino-acid composition after loop deletion drives domain swapping
Protein Sci., 26, 2017
|
|
1NG5
 
 | |
5XJQ
 
 | |
6Y4C
 
 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using an XtalTool support | | 分子名称: | CHLORIDE ION, Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Shilova, A, Hakansson, M, Welin, M, Kovacic, R, Mueller, U, Logan, D.T. | | 登録日 | 2020-02-20 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
1NI9
 
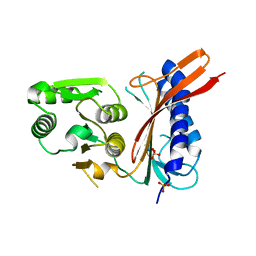 | | 2.0 A structure of glycerol metabolism protein from E. coli | | 分子名称: | Protein glpX, SULFATE ION | | 著者 | Sanishvili, R, Brunzelle, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2002-12-23 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
5XMF
 
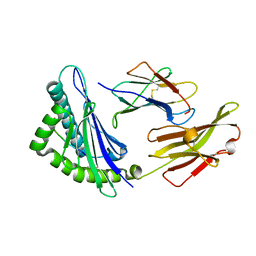 | | Crystal structure of feline MHC class I for 2,1 angstrom | | 分子名称: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | 著者 | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | 登録日 | 2017-05-15 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
6Y9E
 
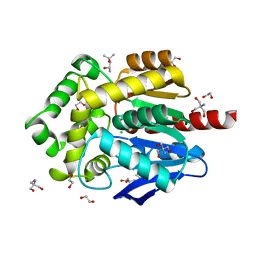 | | Crystal structure of putative ancestral haloalkane dehalogenase AncHLD2 (node 2) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Chaloupkova, R, Damborsky, J, Marek, M. | | 登録日 | 2020-03-09 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structures of hyperstable ancestral haloalkane dehalogenases show restricted conformational dynamics.
Comput Struct Biotechnol J, 18, 2020
|
|
5XQM
 
 | |
6Y7C
 
 | | Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) | | 分子名称: | 20S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Shayan, R, Plassart, L, Plisson-Chastang, C. | | 登録日 | 2020-02-28 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Molecules, 25, 2020
|
|
7NB6
 
 | |
5X6V
 
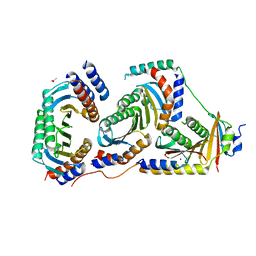 | | Crystal structure of human heteroheptameric complex | | 分子名称: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | 著者 | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | 登録日 | 2017-02-23 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
6YLE
 
 | |
6YMF
 
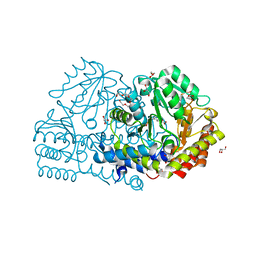 | | Crystal structure of serine hydroxymethyltransferase from Aphanothece halophytica in the PLP-Serine external aldimine state | | 分子名称: | GLYCEROL, PENTAETHYLENE GLYCOL, Serine hydroxymethyltransferase, ... | | 著者 | Ruszkowski, M, Sekula, B, Nogues, I, Tramonti, A, Angelaccio, S, Contestabile, R. | | 登録日 | 2020-04-08 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Structural and kinetic properties of serine hydroxymethyltransferase from the halophytic cyanobacterium Aphanothece halophytica provide a rationale for salt tolerance.
Int.J.Biol.Macromol., 159, 2020
|
|
7N5U
 
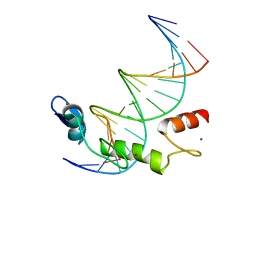 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | 分子名称: | DNA Strain II, DNA Strand I, ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
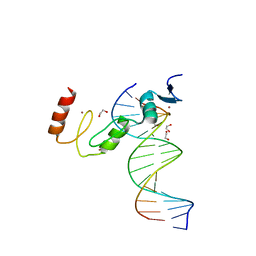 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | 分子名称: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
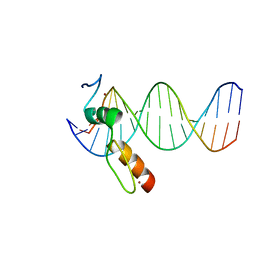 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | 分子名称: | DNA Strand I, DNA Strand II, ZINC ION, ... | | 著者 | Horton, J.R, Ren, R, Cheng, X. | | 登録日 | 2021-06-06 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.08 Å) | | 主引用文献 | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
2N75
 
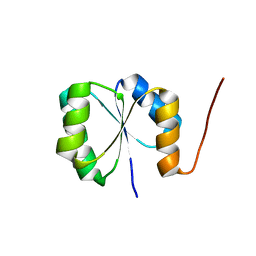 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | 分子名称: | De novo designed protein | | 著者 | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2015-09-03 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2N9H
 
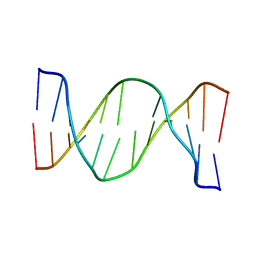 | | Glucose as a nuclease mimic in DNA | | 分子名称: | DNA (5'-D(*CP*TP*AP*GP*CP*(GL6)P*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*TP*GP*CP*TP*AP*G)-3') | | 著者 | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Muro, A, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | 登録日 | 2015-11-25 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5XKR
 
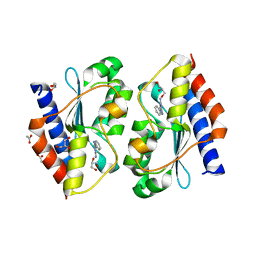 | | Crystal structure of Msmeg3575 in complex with benzoguanamine | | 分子名称: | 6-phenyl-1,3,5-triazine-2,4-diamine, ACETATE ION, CMP/dCMP deaminase, ... | | 著者 | Gaded, V.M, Anand, R. | | 登録日 | 2017-05-09 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
6Y7M
 
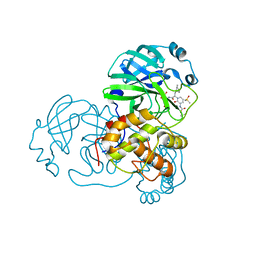 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | Zhang, L, Lin, D, Hilgenfeld, R. | | 登録日 | 2020-03-01 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
