6N9K
 
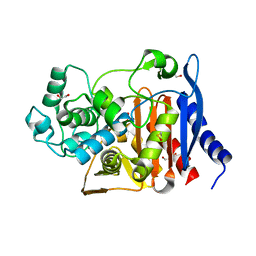 | | Beta-lactamase from Escherichia coli str. Sakai | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | 著者 | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-12-03 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Beta-lactamase from Escherichia coli str. Sakai
to be published
|
|
5YUK
 
 | |
5NE6
 
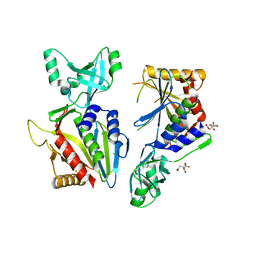 | |
1U9L
 
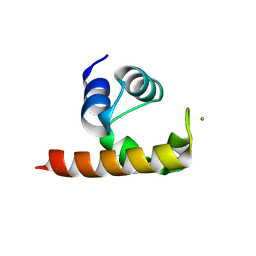 | | Structural basis for a NusA- protein N interaction | | 分子名称: | GOLD ION, Lambda N, Transcription elongation protein nusA | | 著者 | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | 登録日 | 2004-08-10 | | 公開日 | 2004-08-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6MXS
 
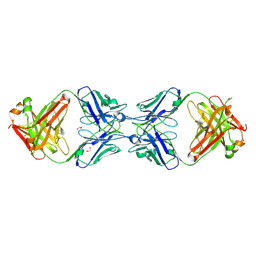 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98F,HC-G99M] | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | 著者 | Shi, R, Picard, M.-E, Manenda, M.S. | | 登録日 | 2018-10-31 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6NAZ
 
 | | Crystal structure of DIRAS 2/3 chimera in complex with GDP | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, GTP-binding protein Di-Ras2,GTP-binding protein Di-Ras3,GTP-binding protein Di-Ras2, ... | | 著者 | Gilbert, Y.H, Reger, A, Sharma, R, Bast, J, Kim, C. | | 登録日 | 2018-12-06 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.081 Å) | | 主引用文献 | Crystal structure of DIRAS 2/3 chimera in complex with GDP
To Be Published
|
|
6NB3
 
 | | MERS-CoV complex with human neutralizing LCA60 antibody Fab fragment (state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | 著者 | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2018-12-06 | | 公開日 | 2019-02-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
4MZW
 
 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | 分子名称: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-09-30 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
5MZZ
 
 | | Crystal structure of the decarboxylase AibA/AibB in complex with 3-methylglutaconate | | 分子名称: | 3-methylpent-2-enedioic acid, ACETATE ION, GLYCEROL, ... | | 著者 | Bock, T, Luxenburger, E, Hoffmann, J, Schuetza, V, Feiler, C, Mueller, R, Blankenfeldt, W. | | 登録日 | 2017-02-02 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | AibA/AibB Induces an Intramolecular Decarboxylation in Isovalerate Biosynthesis by Myxococcus xanthus.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
3RK5
 
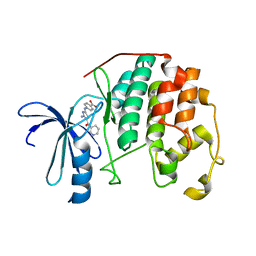 | | CDK2 in complex with inhibitor RC-2-72 | | 分子名称: | 4-{[4-amino-5-(pyridin-3-ylcarbonyl)-1,3-thiazol-2-yl]amino}benzoic acid, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-04-17 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6NF8
 
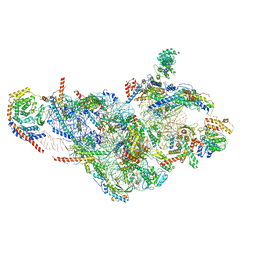 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | 分子名称: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | 著者 | Sharma, M, Koripella, R, Agrawal, R. | | 登録日 | 2018-12-19 | | 公開日 | 2019-02-27 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
3DPF
 
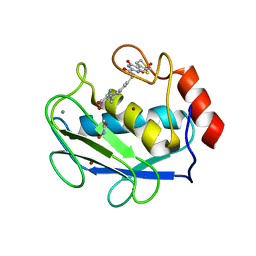 | | Crystal structure of the complex between MMP-8 and a non-zinc chelating inhibitor | | 分子名称: | ACETOHYDROXAMIC ACID, CALCIUM ION, N-{[2-(2-amino-3,4-dioxocyclobut-1-en-1-yl)-1,2,3,4-tetrahydroisoquinolin-7-yl]methyl}-4-oxo-3,5,6,8-tetrahydro-4H-thiopyrano[4',3':4,5]thieno[2,3-d]pyrimidine-2-carboxamide 7,7-dioxide, ... | | 著者 | Pochetti, G, Montanari, R, Mazza, F. | | 登録日 | 2008-07-08 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Extra Binding Region Induced by Non-Zinc Chelating Inhibitors into the S(1)' Subsite of Matrix Metalloproteinase 8 (MMP-8)
J.Med.Chem., 52, 2009
|
|
4J9X
 
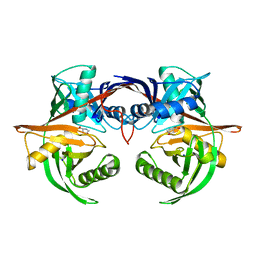 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline | | 分子名称: | 4-HYDROXYPROLINE, Proline racemase family protein, SODIUM ION | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-02-17 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with trans-4-hydroxy-l-proline
To be Published
|
|
3RPR
 
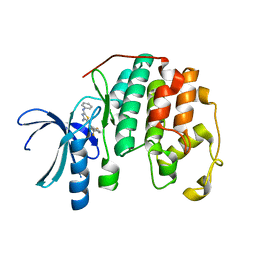 | | CDK2 in complex with inhibitor RC-2-49 | | 分子名称: | Cyclin-dependent kinase 2, [4-amino-2-(phenylamino)-1,3-thiazol-5-yl][3-(trifluoromethyl)phenyl]methanone | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-04-27 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3RPY
 
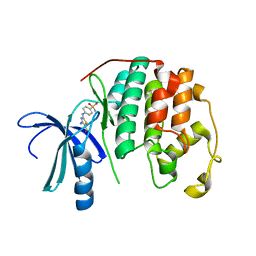 | | CDK2 in complex with inhibitor RC-2-40 | | 分子名称: | 4-amino-2-[(4-sulfamoylphenyl)amino]-1,3-thiazole-5-carboxamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-04-27 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
5NCI
 
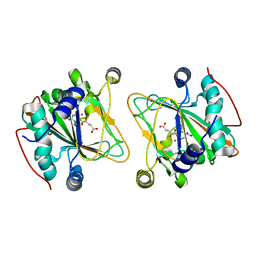 | | GriE in complex with cobalt, alpha-ketoglutarate and l-leucine | | 分子名称: | 2-OXOGLUTARIC ACID, COBALT (II) ION, LEUCINE, ... | | 著者 | Lukat, P, Blankenfeldt, W, Mueller, R. | | 登録日 | 2017-03-05 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.755 Å) | | 主引用文献 | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
5NE7
 
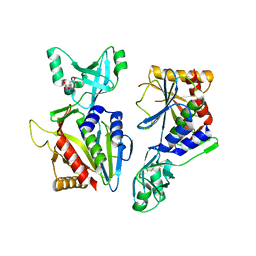 | |
6NKC
 
 | | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome. | | 分子名称: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | 著者 | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | 登録日 | 2019-01-07 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome.
To Be Published
|
|
6NKG
 
 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | 分子名称: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | 著者 | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | 登録日 | 2019-01-07 | | 公開日 | 2020-01-22 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | 分子名称: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | 著者 | Meng, R, Chang, J, Zhang, J. | | 登録日 | 2019-01-10 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
4R15
 
 | | High-resolution crystal structure of Z-DNA in complex with Cr3+ cations | | 分子名称: | CHROMIUM ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | 著者 | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | 登録日 | 2014-08-04 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | High-resolution crystal structure of Z-DNA in complex with Cr(3+) cations.
J.Biol.Inorg.Chem., 20, 2015
|
|
3R8Z
 
 | | CDK2 in complex with inhibitor RC-1-136 | | 分子名称: | Cyclin-dependent kinase 2, [4-amino-2-(prop-2-en-1-ylamino)-1,3-thiazol-5-yl](pyridin-4-yl)methanone | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-24 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
7JUO
 
 | | CBP bromodomain complexed with YF2-23 | | 分子名称: | CREB-binding protein, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(1-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-3-yl)-1H-indol-4-yl}ethanesulfonamide | | 著者 | Ratia, K.M, Xiong, R, Principe, D, Li, Y, Huang, F, Rana, A, Thatcher, G. | | 登録日 | 2020-08-20 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3RAK
 
 | | CDK2 in complex with inhibitor RC-2-32 | | 分子名称: | 4-[(4-amino-5-benzoyl-1,3-thiazol-2-yl)amino]benzenesulfonamide, Cyclin-dependent kinase 2 | | 著者 | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | 登録日 | 2011-03-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
6NEQ
 
 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | 分子名称: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | 著者 | Sharma, M, Koripella, R, Agrawal, R. | | 登録日 | 2018-12-18 | | 公開日 | 2019-02-27 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
