1JZY
 
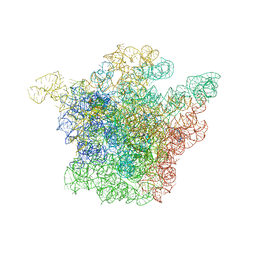 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | 分子名称: | 23S rRNA, ERYTHROMYCIN A, MAGNESIUM ION, ... | | 著者 | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | 登録日 | 2001-09-17 | | 公開日 | 2001-10-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1YLS
 
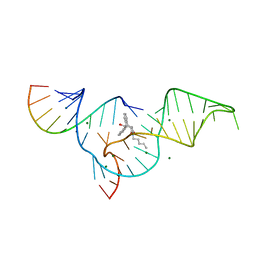 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | 分子名称: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | 著者 | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | 登録日 | 2005-01-19 | | 公開日 | 2005-02-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
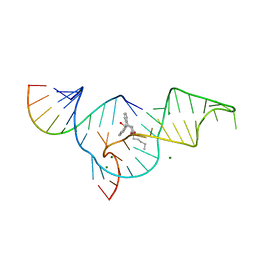 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | 分子名称: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | 著者 | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | 登録日 | 2005-01-18 | | 公開日 | 2005-02-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1OLL
 
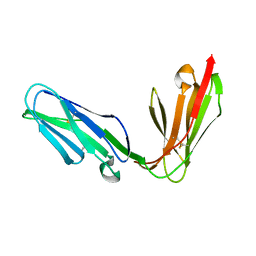 | | Extracellular region of the human receptor NKp46 | | 分子名称: | 1,2-ETHANEDIOL, NK RECEPTOR | | 著者 | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | 登録日 | 2003-08-07 | | 公開日 | 2003-09-04 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1YM7
 
 | | G Protein-Coupled Receptor Kinase 2 (GRK2) | | 分子名称: | Beta-adrenergic receptor kinase 1 | | 著者 | Lodowski, D.T, Barnhill, J.F, Pyskadlo, R.M, Ghirlando, R, Sterne-Marr, R, Tesmer, J.J.G. | | 登録日 | 2005-01-20 | | 公開日 | 2005-07-05 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | The role of Gbetagamma and domain interfaces in the activation of G protein-coupled receptor kinase 2
Biochemistry, 44, 2005
|
|
1ID4
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC SITE MUTANT (H157Q) OF THE HUMAN CYTOMEGALOVIRUS PROTEASE | | 分子名称: | CAPSID PROTEIN P40: ASSEMBLIN PROTEASE | | 著者 | Khayat, R, Batra, R, Massariol, M.J, Lagace, L, Tong, L. | | 登録日 | 2001-04-03 | | 公開日 | 2001-06-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Investigating the role of histidine 157 in the catalytic activity of human cytomegalovirus protease.
Biochemistry, 40, 2001
|
|
5NM2
 
 | | A2A Adenosine receptor cryo structure | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | 著者 | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | 登録日 | 2017-04-05 | | 公開日 | 2017-09-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.948 Å) | | 主引用文献 | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
1AVR
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF HUMAN ANNEXIN V AFTER REFINEMENT. IMPLICATIONS FOR STRUCTURE, MEMBRANE BINDING AND ION CHANNEL FORMATION OF THE ANNEXIN FAMILY OF PROTEINS | | 分子名称: | ANNEXIN V, CALCIUM ION, SULFATE ION | | 著者 | Huber, R, Berendes, R, Burger, A, Schneider, M, Karshikov, A, Luecke, H, Roemisch, J, Paques, E. | | 登録日 | 1991-10-17 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
4NHO
 
 | | Structure of the spliceosomal DEAD-box protein Prp28 | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, MERCURY (II) ION, ... | | 著者 | Moehlmann, S, Neumann, P, Ficner, R. | | 登録日 | 2013-11-05 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5LY2
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Macrocyclic PEPTIDE Inhibitor CP2_R6Kme3 (13-mer) | | 分子名称: | CHLORIDE ION, CP2_R6Kme3, GLYCEROL, ... | | 著者 | Chowdhury, R, Madden, S.K, Hopkinson, R, Schofield, C.J. | | 登録日 | 2016-09-23 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Highly selective inhibition of histone demethylases by de novo macrocyclic peptides.
Nat Commun, 8, 2017
|
|
5K7H
 
 | | Crystal structure of AibR in complex with the effector molecule isovaleryl coenzyme A | | 分子名称: | CHLORIDE ION, Isovaleryl-coenzyme A, NICKEL (II) ION, ... | | 著者 | Bock, T, Volz, C, Mueller, R, Blankenfeldt, W. | | 登録日 | 2016-05-26 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The AibR-isovaleryl coenzyme A regulator and its DNA binding site - a model for the regulation of alternative de novo isovaleryl coenzyme A biosynthesis in Myxococcus xanthus.
Nucleic Acids Res., 45, 2017
|
|
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | 分子名称: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | 著者 | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-02-04 | | 公開日 | 2013-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
4J6F
 
 | | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230 | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative alcohol dehydrogenase | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-02-11 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of putative alcohol dehydrogenase from Sinorhizobium meliloti 1021, NYSGRC-Target 012230
To be Published
|
|
1Z5H
 
 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | 分子名称: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | 著者 | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | 登録日 | 2005-03-18 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
1NZJ
 
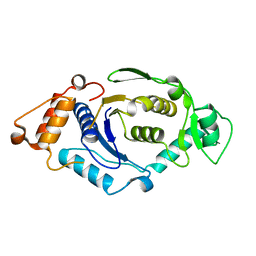 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | 分子名称: | Hypothetical protein yadB, ZINC ION | | 著者 | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | 登録日 | 2003-02-18 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
1K01
 
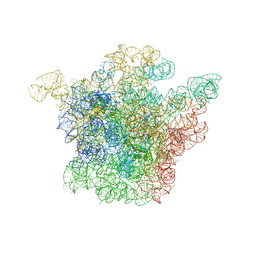 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | 分子名称: | 23S rRNA, CHLORAMPHENICOL, MAGNESIUM ION, ... | | 著者 | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | 登録日 | 2001-09-17 | | 公開日 | 2001-10-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1YWI
 
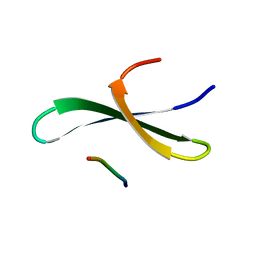 | | Structure of the FBP11WW1 domain complexed to the peptide APPTPPPLPP | | 分子名称: | Formin, Formin-binding protein 3 | | 著者 | Pires, J.R, Parthier, C, Aido-Machado, R, Wiedemann, U, Otte, L, Boehm, G, Rudolph, R, Oschkinat, H. | | 登録日 | 2005-02-18 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain.
J.Mol.Biol., 348, 2005
|
|
1JZX
 
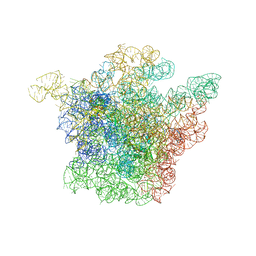 | | Structural Basis for the Interaction of Antibiotics with the Peptidyl Transferase Center in Eubacteria | | 分子名称: | 23S rRNA, CLINDAMYCIN, MAGNESIUM ION, ... | | 著者 | Schluenzen, F, Zarivach, R, Harms, J, Bashan, A, Tocilj, A, Albrecht, R, Yonath, A, Franceschi, F. | | 登録日 | 2001-09-17 | | 公開日 | 2001-10-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria.
Nature, 413, 2001
|
|
1XFQ
 
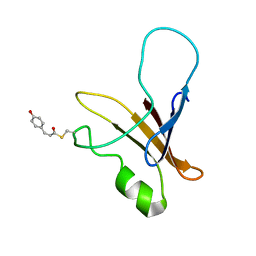 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | 登録日 | 2004-09-15 | | 公開日 | 2005-08-16 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1XGV
 
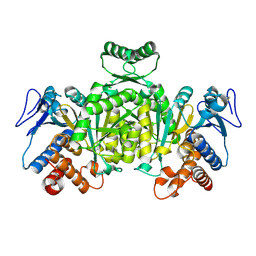 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix | | 分子名称: | Isocitrate dehydrogenase | | 著者 | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N.-K, Ladenstein, R. | | 登録日 | 2004-09-17 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
1RBG
 
 | |
1NJO
 
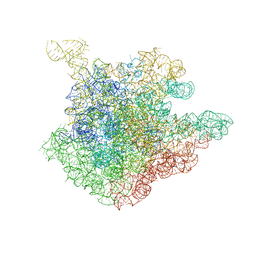 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a short substrate analog ACCPuromycin (ACCP) | | 分子名称: | 23S ribosomal RNA, RNA ACC(Puromycin) | | 著者 | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | 登録日 | 2003-01-02 | | 公開日 | 2003-02-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NKM
 
 | | Complex structure of HCMV Protease and a peptidomimetic inhibitor | | 分子名称: | Assemblin, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | 著者 | Khayat, R, Batra, R, Qian, C, Halmos, T, Bailey, M, Tong, L. | | 登録日 | 2003-01-03 | | 公開日 | 2003-02-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and Biochemical Studies of Inhibitor Binding to Human Cytomegalovirus Protease
Biochemistry, 42, 2003
|
|
1XHS
 
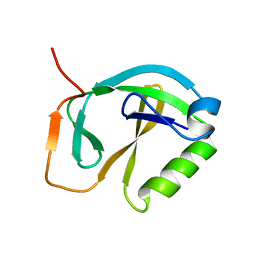 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | 分子名称: | Hypothetical UPF0131 protein ytfP | | 著者 | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2004-09-20 | | 公開日 | 2005-01-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
6X4I
 
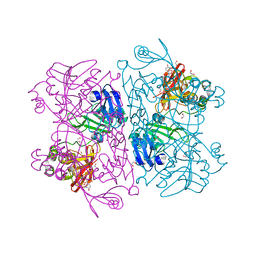 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | 分子名称: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | 著者 | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
