1WYT
 
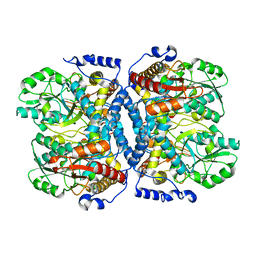 | | Crystal structure of glycine decarboxylase (P-protein) of the glycine cleavage system, in apo form | | Descriptor: | glycine dehydrogenase (decarboxylating) subunit 1, glycine dehydrogenase subunit 2 (P-protein) | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of P-protein of the glycine cleavage system: implications for nonketotic hyperglycinemia
Embo J., 24, 2005
|
|
6E2R
 
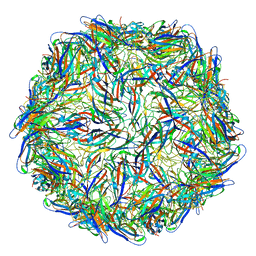 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
1NEK
 
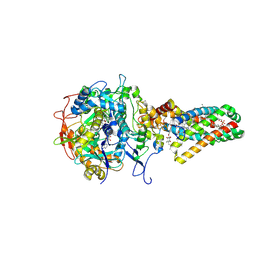 | | Complex II (Succinate Dehydrogenase) From E. Coli with ubiquinone bound | | Descriptor: | CALCIUM ION, CARDIOLIPIN, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Yankovskaya, V, Horsefield, R, Tornroth, S, Luna-Chavez, C, Miyoshi, H, Leger, C, Byrne, B, Cecchini, G, Iwata, S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Architecture of succinate dehydrogenase and
reactive oxygen species generation.
Science, 299, 2003
|
|
1NJQ
 
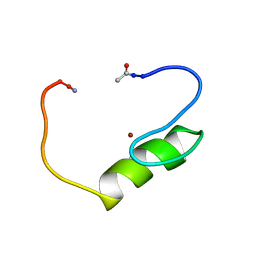 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
1NAV
 
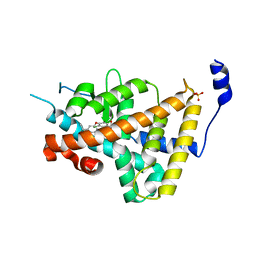 | | Thyroid Receptor Alpha in complex with an agonist selective for Thyroid Receptor Beta1 | | Descriptor: | SULFATE ION, hormone receptor alpha 1, THRA1, ... | | Authors: | Ye, L, Li, Y.L, Mellstrom, K, Mellin, C, Bladh, L.G, Koehler, K, Garg, N, Garcia Collazo, A.M, Litten, C, Husman, B, Persson, K, Ljunggren, J, Grover, G, Sleph, P.G, George, R, Malm, J. | | Deposit date: | 2002-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thyroid receptor ligands. 1. Agonist ligands selective for the thyroid receptor beta1.
J.Med.Chem., 46, 2003
|
|
1NBK
 
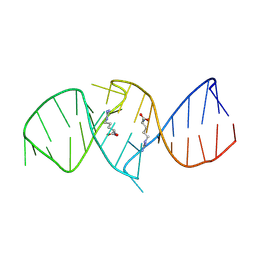 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
1WKB
 
 | |
6EEA
 
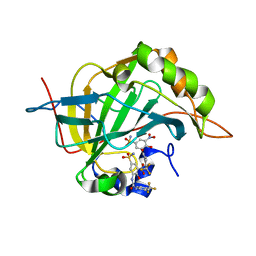 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
1WNE
 
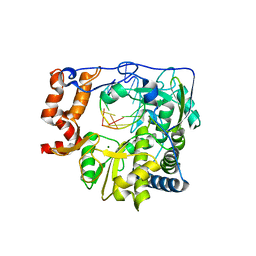 | | Foot and Mouth Disease Virus RNA-dependent RNA polymerase in complex with a template-primer RNA | | Descriptor: | 5'-R(*CP*AP*UP*GP*GP*GP*CP*C)-3', 5'-R(*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2004-07-31 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Foot-and-Mouth Disease Virus RNA-dependent RNA Polymerase and Its Complex with a Template-Primer RNA
J.Biol.Chem., 279, 2004
|
|
6EGE
 
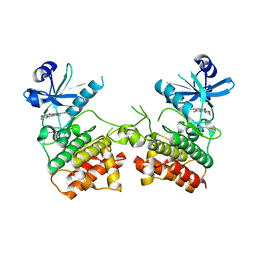 | |
6EID
 
 | | Crystal structure of wild-type Channelrhodopsin 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, PHOSPHATE ION, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | Deposit date: | 2017-09-19 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
1WP6
 
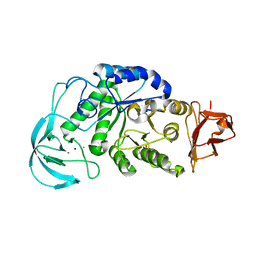 | | Crystal structure of maltohexaose-producing amylase from alkalophilic Bacillus sp.707. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glucan 1,4-alpha-maltohexaosidase, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-08-31 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
1WPX
 
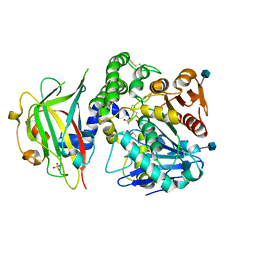 | | Crystal structure of carboxypeptidase Y inhibitor complexed with the cognate proteinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase Y, Carboxypeptidase Y inhibitor, ... | | Authors: | Mima, J, Hayashida, M, Fujii, T, Narita, Y, Hayashi, R, Ueda, M, Hata, Y. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the carboxypeptidase y inhibitor i(c) in complex with the cognate proteinase reveals a novel mode of the proteinase-protein inhibitor interaction
J.Mol.Biol., 346, 2005
|
|
1NEN
 
 | | Complex II (Succinate Dehydrogenase) From E. Coli with Dinitrophenol-17 inhibitor co-crystallized at the ubiquinone binding site | | Descriptor: | 2-[1-METHYLHEXYL]-4,6-DINITROPHENOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Yankovskaya, V, Horsefield, R, Tornroth, S, Luna-Chavez, C, Miyoshi, H, Leger, C, Byrne, B, Cecchini, G, Iwata, S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Architecture of succinate dehydrogenase and
reactive oxygen species generation
Science, 299, 2003
|
|
1WPP
 
 | | Structure of Streptococcus gordonii inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Probable manganese-dependent inorganic pyrophosphatase, SULFATE ION, ... | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1NGO
 
 | | NMR Structure of Putative 3' Terminator for B. Anthracis pagA Gene Coding Strand | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*TP*TP*GP*TP*AP*AP*GP*AP*AP*AP*TP*AP*CP*AP*AP*GP*GP*AP*GP*AP*G)-3' | | Authors: | Shiflett, P.R, Taylor-McCabe, K.J, Michalczyk, R, Silks, L.A, Gupta, G. | | Deposit date: | 2002-12-17 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on the Hairpins at the 3' Untranslated Region of an Anthrax Toxin Gene
Biochemistry, 42, 2003
|
|
1WQ3
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6EPC
 
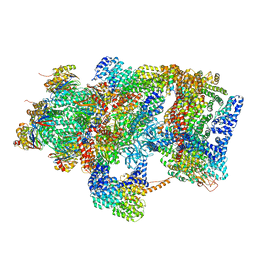 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
1WTJ
 
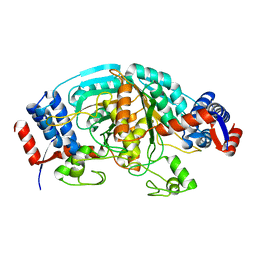 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | Descriptor: | ureidoglycolate dehydrogenase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
4HOV
 
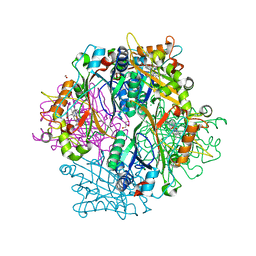 | | DypB N246A in complex with manganese | | Descriptor: | CHLORIDE ION, DypB, FORMIC ACID, ... | | Authors: | Grigg, J.C, Singh, R, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved Manganese-Oxidizing Activity of DypB, a Peroxidase from a Lignolytic Bacterium.
Acs Chem.Biol., 8, 2013
|
|
1NKO
 
 | | Energetic and structural basis of sialylated oligosaccharide recognition by the natural killer cell inhibitory receptor p75/AIRM1 or Siglec-7 | | Descriptor: | Sialic acid binding Ig-like lectin 7 | | Authors: | Dimasi, N, Attril, H, van Aalten, D.M.F, Moretta, L, Biassoni, R, Mariuzza, R.A. | | Deposit date: | 2003-01-03 | | Release date: | 2003-04-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the saccharide-binding domain of the human natural killer cell inhibitory receptor p75/AIRM1.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1NDQ
 
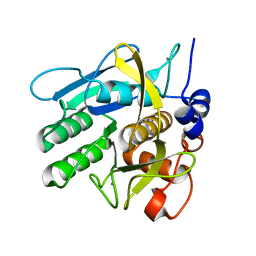 | | Bacillus lentus subtilisin | | Descriptor: | CALCIUM ION, Subtilisin Savinase | | Authors: | Pan, X, Bott, R, Glatz, C.E. | | Deposit date: | 2002-12-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Subtilisin surface properties and crystal growth kinetics
J.CRYST.GROWTH, 254, 2003
|
|
1WD5
 
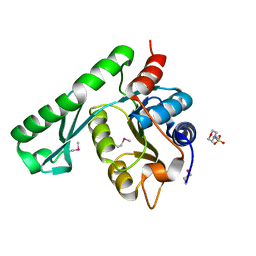 | | Crystal structure of TT1426 from Thermus thermophilus HB8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, hypothetical protein TT1426 | | Authors: | Shibata, R, Kukimoto-Niino, M, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-11 | | Release date: | 2004-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution
Protein Sci., 14, 2005
|
|
1N9J
 
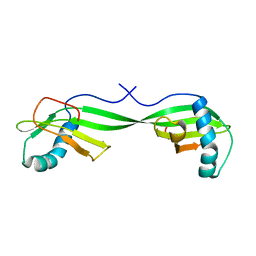 | | Solution Structure of the 3D domain swapped dimer of Stefin A | | Descriptor: | Cystatin A | | Authors: | Staniforth, R.A, Giannini, S, Higgins, L.D, Conroy, M.J, Hounslow, A.M, Jerala, R, Craven, C.J, Waltho, J.P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional domain swapping in the folded and molten-globule states of cystatins, an amyloid-forming structural superfamily
Embo J., 20, 2001
|
|
1WXR
 
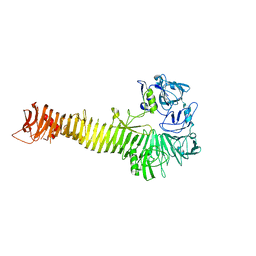 | | Crystal structure of Heme Binding protein, an autotransporter hemoglobine protease from pathogenic Escherichia coli | | Descriptor: | haemoglobin protease | | Authors: | Otto, B.R, Sijbrandi, R, Luirink, J, Oudega, B, Heddle, J.G, Mizutani, K, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-01-31 | | Release date: | 2005-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of heme binding protein, an autotransporter hemoglobin protease from pathogenic escherichia coli
J.Biol.Chem., 280, 2005
|
|
