3SAL
 
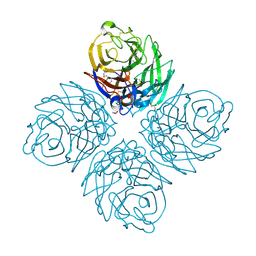 | | Crystal Structure of Influenza A Virus Neuraminidase N5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, M.Y, Qi, J.X, Liu, Y, Vavricka, C.J, Wu, Y, Li, Q, Gao, G.F. | | Deposit date: | 2011-06-03 | | Release date: | 2011-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza a virus n5 neuraminidase has an extended 150-cavity
J.Virol., 85, 2011
|
|
3UYW
 
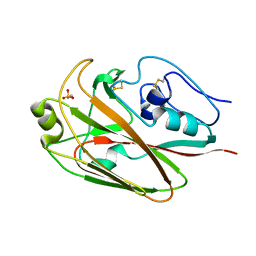 | | Crystal structures of globular head of 2009 pandemic H1N1 hemagglutinin | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Hemagglutinin | | Authors: | Xuan, C.L, Shi, Y, Qi, J.X, Xiao, H.X, Gao, G.F. | | Deposit date: | 2011-12-06 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines
Protein Cell, 2, 2011
|
|
7XA7
 
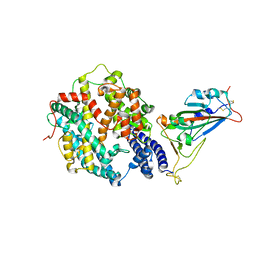 | | Crystal structure of SARS-CoV-2 receptor-binding domain in complex with intermediate horseshoe bat ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Tang, L.F, Zhang, D, Han, P, Qi, J.X. | | Deposit date: | 2022-03-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of SARS-CoV-2 and its variants binding to intermediate horseshoe bat ACE2.
Int J Biol Sci, 18, 2022
|
|
7XBG
 
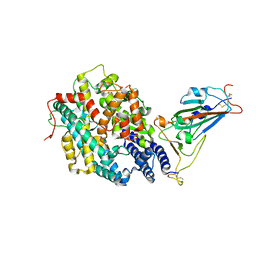 | | The crystal structure of RshSTT182/200 RBD-insert2-T346R-Y496G mutant in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBH
 
 | | The complex structure of RshSTT182/200 RBD bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7XBF
 
 | | The complex structure of RshSTT182/200 RBD-insert2 bound to human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, RshSTT182/200 coronavirus receptor binding domain insert2, ... | | Authors: | Hu, Y, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Host range and structural analysis of bat-origin RshSTT182/200 coronavirus binding to human ACE2 and its animal orthologs.
Embo J., 42, 2023
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-26 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
4PBP
 
 | | crystal structure of zebrafish short-chain pentraxin protein | | Descriptor: | C-reactive protein, CALCIUM ION, GLYCEROL | | Authors: | Chen, R, Qi, J.X, George, F.G, Xia, C. | | Deposit date: | 2014-04-13 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structures for short-chain pentraxin from zebrafish demonstrate a cyclic trimer with new recognition and effector faces.
J.Struct.Biol., 189, 2015
|
|
3Q2C
 
 | | Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 3 | | Authors: | Ryu, M, Chen, Y, Qi, J.X, Liu, J, Shi, Y, Cheng, H, Gao, G.F. | | Deposit date: | 2010-12-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence
Plos One, 6, 2011
|
|
5GS6
 
 | | Full-length NS1 structure of Zika virus from 2015 Brazil strain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NS1 of Zika virus from 2015 Brazil strain | | Authors: | Xu, X.Y, Song, H, Qi, J.X, Shi, Y, Gao, G.F. | | Deposit date: | 2016-08-14 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure
Embo J., 35, 2016
|
|
4PBO
 
 | |
9IK2
 
 | | The co-crystal structure of SARS-CoV-2 Mpro in complex with compound H109 | | Descriptor: | 3C-like proteinase, tert-butyl N-[(2S)-1-[[(2S)-1-[[(2S)-1-azanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]carbamate | | Authors: | Feng, Y, Zheng, W.Y, Han, P, Fu, L.F, Qi, J.X. | | Deposit date: | 2024-06-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided discovery of a small molecule inhibitor of SARS-CoV-2 main protease with potent in vitro and in vivo antiviral activities
To Be Published
|
|
7YJ3
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7BV0
 
 | | HEV-C E2s | | Descriptor: | Protein ORF2 | | Authors: | Bai, C.Z, Qi, J.X. | | Deposit date: | 2020-04-08 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of emerging human-infected hepatitis E virus E2s at 1.8 Angstroms resolution
To Be Published
|
|
4FVK
 
 | | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Sun, X.M, Li, Z.X, Liu, Y, Vavricka, C.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
