9ERO
 
 | |
9ERM
 
 | |
9ERN
 
 | |
9EO9
 
 | | SF type tau filament from V337M mutant | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Qi, C, Scheres, S.H.W, Michel, G. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tau filaments with the Alzheimer fold in human MAPT mutants V337M and R406W.
Nat.Struct.Mol.Biol., 2025
|
|
9EO7
 
 | | PHF type tau filament from V337M mutant | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Qi, C, Scheres, S.H.W, Michel, G. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tau filaments with the Alzheimer fold in human MAPT mutants V337M and R406W.
Nat.Struct.Mol.Biol., 2025
|
|
9EOE
 
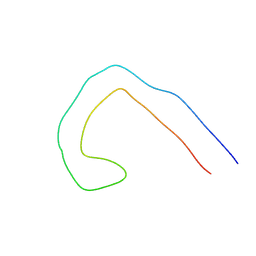 | | TF type tau filament from V337M mutant | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau | | Authors: | Qi, C, Scheres, S.H.W, Michel, G. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-10 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Tau filaments with the Alzheimer fold in human MAPT mutants V337M and R406W.
Nat.Struct.Mol.Biol., 2025
|
|
9EOG
 
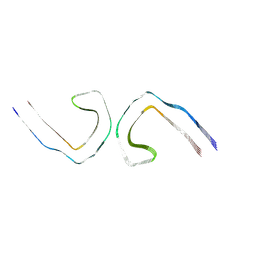 | |
9EOH
 
 | |
6R4O
 
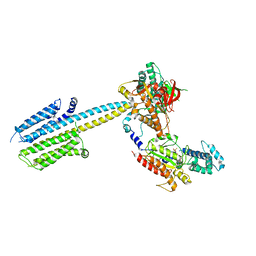 | | Structure of a truncated adenylyl cyclase bound to MANT-GTP, forskolin and an activated stimulatory Galphas protein | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase 9, ... | | Authors: | Qi, C, Sorrentino, S, Medalia, O, Korkhov, V.M. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structure of a membrane adenylyl cyclase bound to an activated stimulatory G protein.
Science, 364, 2019
|
|
8CAX
 
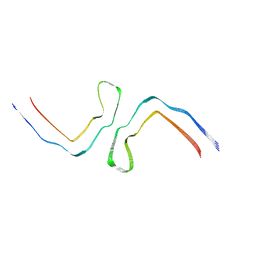 | | Structure of Tau filaments Type II from Subacute Sclerosing Panencephalitis | | Descriptor: | Microtubule-associated protein tau | | Authors: | Qi, C, Hasegawa, M, Takao, M, Sakai, M, Akagi, M, Iwasaki, Y, Yoshida, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Identical tau filaments in subacute sclerosing panencephalitis and chronic traumatic encephalopathy.
Acta Neuropathol Commun, 11, 2023
|
|
8CAQ
 
 | | Structure of Tau filaments Type I from Subacute Sclerosing Panencephalitis | | Descriptor: | Microtubule-associated protein tau | | Authors: | Qi, C, Hasegawa, M, Takao, M, Sakai, M, Akagi, M, Iwasaki, Y, Yoshida, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-01-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Identical tau filaments in subacute sclerosing panencephalitis and chronic traumatic encephalopathy.
Acta Neuropathol Commun, 11, 2023
|
|
7PDH
 
 | |
7PD4
 
 | |
7PD8
 
 | |
7PDD
 
 | |
7PDG
 
 | |
7PDF
 
 | |
7PDE
 
 | |
7Z1T
 
 | |
7Z22
 
 | |
7ZXO
 
 | |
7ZXQ
 
 | |
7ZXP
 
 | |
7ZXM
 
 | |
7ZXT
 
 | |
