2P66
 
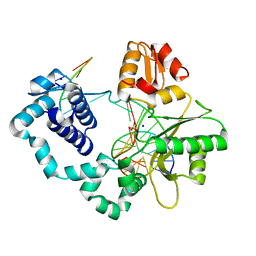 | | Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), DNA (5'-D(P*(3DR)P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Prasad, R, Batra, V.K, Yang, X.-P, Krahn, J.M, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the DNA Polymerase beta deoxyribose phosphate lyase mechanism
DNA REPAIR, 4, 2005
|
|
4XZQ
 
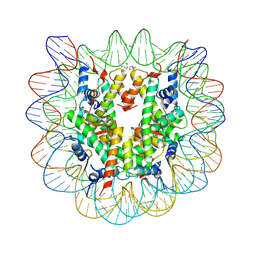 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-02-04 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
8U1T
 
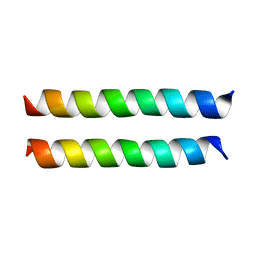 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
1DK2
 
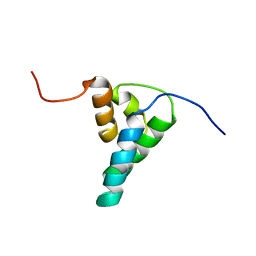 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
1DK3
 
 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
3MBY
 
 | | Ternary complex of DNA Polymerase BETA with template base A and 8oxodGTP in the active site with a dideoxy terminated primer | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Hou, E.W, Pedersen, L.C, Prasad, R, Wilson, S.H. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4YS3
 
 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
4GFJ
 
 | | Crystal structure of Topo-78, an N-terminal 78kDa fragment of topoisomerase V | | Descriptor: | GLYCEROL, Topoisomerase V, ZINC ION | | Authors: | Rajan, R, Prasad, R, Taneja, B, Wilson, S.H, Mondragon, A. | | Deposit date: | 2012-08-03 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Identification of one of the apurinic/apyrimidinic lyase active sites of topoisomerase V by structural and functional studies.
Nucleic Acids Res., 41, 2013
|
|
4Z66
 
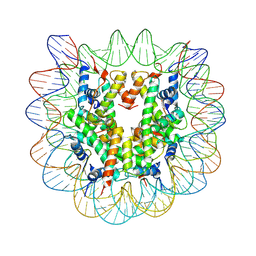 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-04-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
1BNO
 
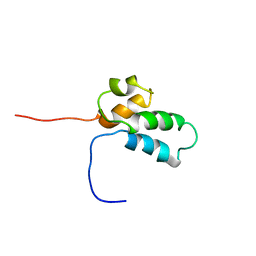 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BNP
 
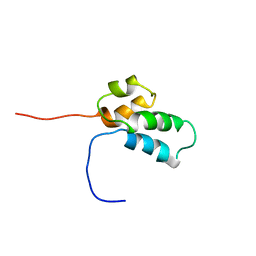 | | NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Liu, D.-J, Prasad, R, Wilson, S.H, Derose, E.F, Mullen, G.P. | | Deposit date: | 1996-04-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal domain of DNA polymerase beta and mapping of the ssDNA interaction interface.
Biochemistry, 35, 1996
|
|
1BPZ
 
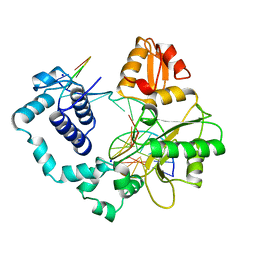 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
1BPY
 
 | | HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*DOC)-3'), ... | | Authors: | Sawaya, M.R, Pelletier, H, Prasad, R, Wilson, S.H, Kraut, J. | | Deposit date: | 1997-04-15 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
1BPX
 
 | | DNA POLYMERASE BETA/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Sawaya, M.R, Prasad, R, Wilson, S.H, Kraut, J, Pelletier, H. | | Deposit date: | 1997-04-11 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism.
Biochemistry, 36, 1997
|
|
4O9M
 
 | | Human DNA polymerase beta complexed with adenylated tetrahydrofuran (abasic site) containing DNA | | Descriptor: | (5'-D(P*GP*TP*CP*GP*G)-3');, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Caglayan, M, Batra, V.K, Sassa, A, Prasad, R, Wilson, S.H. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair.
Nat.Struct.Mol.Biol., 21, 2014
|
|
