4P1V
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with H-type 2 HBGA | | Descriptor: | P domain of VPI, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5497 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
5VX9
 
 | | VP8* of P[6] Human Rotavirus RV3 in complex with LNFP1 | | Descriptor: | Outer capsid protein VP4, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
4F6Z
 
 | | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-b-lactamase active site | | Descriptor: | Beta-lactamase, CITRATE ANION, ZINC ION | | Authors: | Horton, L.B, Shanker, S, Sankaran, B, Mikulski, R, Brown, N.G, Phillips, K, Lykissa, E, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2012-05-15 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-beta-lactamase active site
Antimicrob.Agents Chemother., 56, 2012
|
|
4OPH
 
 | | X-ray structure of full-length H6N6 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Carrillo, B, Choi, J.M, Bornholdt, Z.A, Sankaran, S, Rice, A.P, Prasad, B.V.V. | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.158 Å) | | Cite: | The Influenza A Virus Protein NS1 Displays Structural Polymorphism.
J.Virol., 88, 2014
|
|
4FKD
 
 | | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta | | Descriptor: | Protein kinase C theta type, ZINC ION | | Authors: | Rahman, G.M, Shanker, S, Lewin, N.E, Prasad, B.V.V, Blumberg, P.M, Das, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.633 Å) | | Cite: | Identification of the Activator Binding Residues in the Second Cysteine-Rich Regulatory Domain of Protein Kinase C Theta.
Biochem.J., 451, 2013
|
|
4P3I
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeA HBGA. | | Descriptor: | P domain of VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6941 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P12
 
 | | Native Structure of the P domain from a GI.7 Norovirus variant. | | Descriptor: | Major capsid protein | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-02-24 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6011 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P25
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeY HBGA. | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-01 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4991 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
4P2N
 
 | | Structure of the P domain from a GI.7 Norovirus variant in complex with LeX HBGA | | Descriptor: | Major capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Shanker, S, Czako, R, Sankaran, B, Atmar, R, Estes, M, Prasad, B.V.V. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of determinants of histo-blood group antigen binding specificity in genogroup I noroviruses.
J.Virol., 88, 2014
|
|
5VX4
 
 | | VP8* of a G2P[4] Human Rotavirus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Outer capsid protein VP4 | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VX8
 
 | | VP8* of P[6] Human Rotavirus RV3 | | Descriptor: | Outer capsid protein VP4, SULFATE ION | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
5VX5
 
 | | VP8* of a G2P[4] Human Rotavirus in complex with LNFP1 | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hu, L, Venkataram Prasad, B.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.285 Å) | | Cite: | Glycan recognition in globally dominant human rotaviruses.
Nat Commun, 9, 2018
|
|
4G0J
 
 | |
4I3K
 
 | | Crystal structure of a metabolic reductase with 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one | | Descriptor: | 1-hydroxy-6-(4-hydroxybenzyl)-4-methylpyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, B, Yao, Y, Liu, Z, Deng, L, Anglin, J.L, Jiang, H, Prasad, B.V.V, Song, Y. | | Deposit date: | 2012-11-26 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3056 Å) | | Cite: | Crystallographic Investigation and Selective Inhibition of Mutant Isocitrate Dehydrogenase.
ACS Med Chem Lett, 4, 2013
|
|
2FYQ
 
 | |
2GE8
 
 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2FYR
 
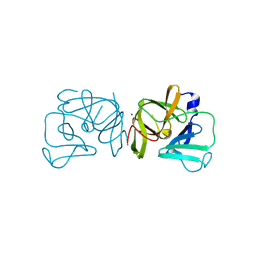 | |
2GEC
 
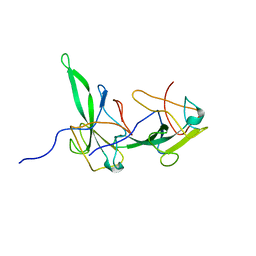 | | Structure of the N-terminal domain of avian infectious bronchitis virus nucleocapsid protein (strain Gray) in a novel dimeric arrangement | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-19 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GE7
 
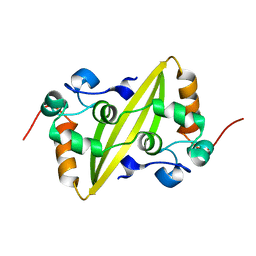 | | Structure of the C-terminal dimerization domain of infectious bronchitis virus nucleocapsid protein | | Descriptor: | Nucleocapsid protein | | Authors: | Jayaram, H, Fan, H, Bowman, B.R, Ooi, A, Jayaram, J, Collisson, E.W, Lescar, J, Prasad, B.V. | | Deposit date: | 2006-03-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation.
J.Virol., 80, 2006
|
|
2GU0
 
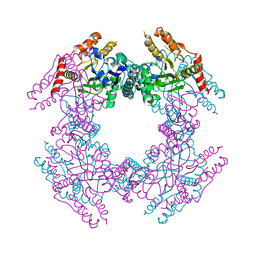 | |
2GX9
 
 | |
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
