8CX6
 
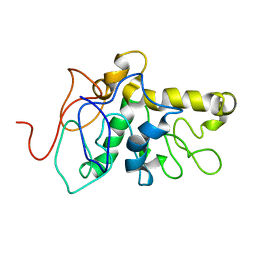 | | TPX2 Minimal Active Domain on Microtubules | | Descriptor: | Targeting protein for Xklp2-A | | Authors: | Guo, C, Alfaro-Aco, R, Russell, R, Zhang, C, Petry, S, Polenova, T. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of protein condensation on microtubules underlying branching microtubule nucleation.
Nat Commun, 14, 2023
|
|
6WAP
 
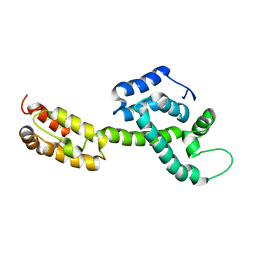 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7M0G
 
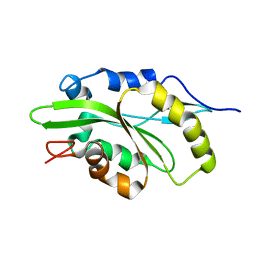 | |
7U8K
 
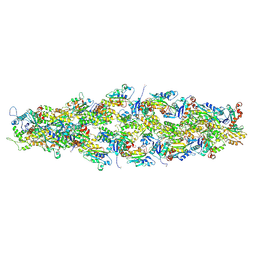 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
2M02
 
 | | 3D structure of cap-gly domain of mammalian dynactin determined by magic angle spinning NMR spectroscopy | | Descriptor: | Dynactin subunit 1 | | Authors: | Yan, S, Hou, G, Schwieters, C.D, Ahmed, S, Williams, J.C, Polenova, T. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Three-Dimensional Structure of CAP-Gly Domain of Mammalian Dynactin Determined by Magic Angle Spinning NMR Spectroscopy: Conformational Plasticity and Interactions with End-Binding Protein EB1.
J.Mol.Biol., 425, 2013
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7SD4
 
 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | Descriptor: | Nucleoprotein | | Authors: | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
2MPX
 
 | |
7RIK
 
 | | Magic-Angle-Spinning NMR Structure of Kinesin-1 Motor Domain Assembled with Microtubules | | Descriptor: | Kinesin-1 heavy chain | | Authors: | Zhang, C, Guo, C, Russell, R.W, Quinn, C.M, Li, M, Williams, J.C, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Magic-angle-spinning NMR structure of the kinesin-1 motor domain assembled with microtubules reveals the elusive neck linker orientation
Nat Commun, 13, 2022
|
|
