5VIK
 
 | |
8EUW
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUU
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUV
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
3H1R
 
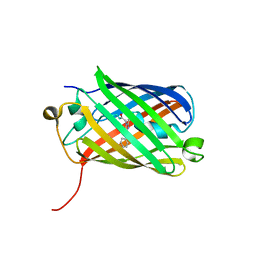 | | Order-disorder structure of fluorescent protein FP480 | | Descriptor: | Fluorescent protein FP480 | | Authors: | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | Deposit date: | 2009-04-13 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3H1O
 
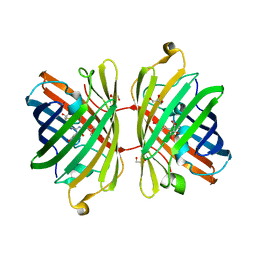 | | The Structure of Fluorescent Protein FP480 | | Descriptor: | Fluorescent protein FP480, GLYCEROL | | Authors: | Pletnev, S, Morozova, K.S, Verkhusha, V.V, Dauter, Z. | | Deposit date: | 2009-04-13 | | Release date: | 2009-09-08 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rotational order-disorder structure of fluorescent protein FP480
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6MGH
 
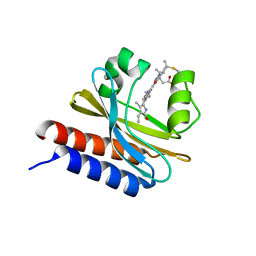 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
4KPI
 
 | |
4Q7R
 
 | |
8DEF
 
 | | Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW24 fab | | Descriptor: | SKW24 Fab heavy chain, SKW24 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Pletnev, S, Tsybovsky, Y, Verardi, R, Roedeger, M, Kwong, P.D. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
3LF3
 
 | | Crystal Structure of Fast Fluorescent Timer Fast-FT | | Descriptor: | Fast Fluorescent Timer Fast-FT | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
3U8A
 
 | |
8DEC
 
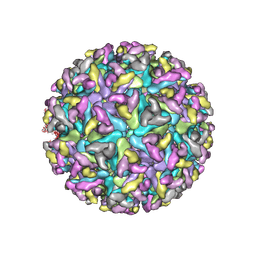 | | Cryo-EM Structure of Western Equine Encephalitis Virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DED
 
 | | Cryo-EM Structure of Western Equine Encephalitis Virus VLP in complex with SKW19 fab | | Descriptor: | SKW19 Fab heavy chain, SKW19 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Pletnev, S, Tsybovsky, Y, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DEE
 
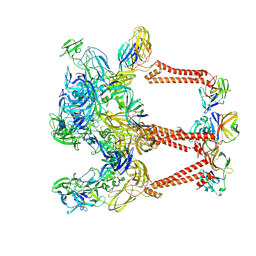 | | Asymmetric Unit of Western Equine Encephalitis Virus | | Descriptor: | Capsid protein, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-06-20 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
8DWO
 
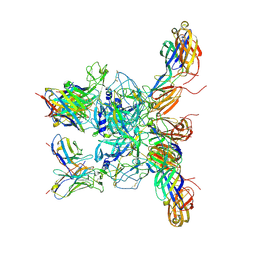 | | Cryo-EM Structure of Eastern Equine Encephalitis Virus in complex with SKE26 Fab | | Descriptor: | Envelope glycoprotein E1, Envelope glycoprotein E2, SKE26 Fab Heavy Chain, ... | | Authors: | Pletnev, S, Verardi, R, Roedeger, M, Kwong, P. | | Deposit date: | 2022-08-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
3LF4
 
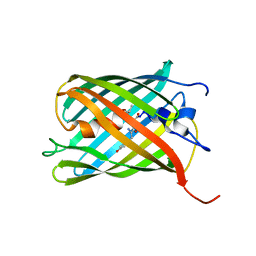 | | Crystal Structure of Fluorescent Timer Precursor Blue102 | | Descriptor: | Fluorescent Timer Precursor Blue102 | | Authors: | Pletnev, S, Dauter, Z. | | Deposit date: | 2010-01-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Understanding blue-to-red conversion in monomeric fluorescent timers and hydrolytic degradation of their chromophores
J.Am.Chem.Soc., 132, 2010
|
|
8DA0
 
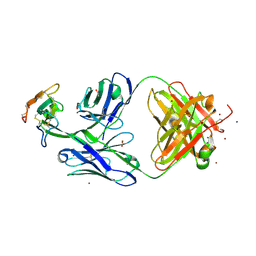 | | Crystal structure of Mamba alpha-neurotoxin in complex with Centi-3FTX-D09 antibody | | Descriptor: | ACETATE ION, Alpha-elapitoxin-Dpp2d, Centi-3FTX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
8D9Y
 
 | | Crystal structure of Taipan alpha-neurotoxin in complex with Centi-3FTX-D09 antibody | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Centi-3FTX-D09 Fab heavy chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
8DA1
 
 | | Crystal structure of Krait alpha-neurotoxin in complex with Centi-3FTX-D09 antibody | | Descriptor: | Alpha-bungarotoxin, Centi-3FTX-D09 Fab heavy chain, Centi-3FTX-D09 Fab light chain | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
8D9Z
 
 | | Crystal structure of Cobra alpha-neurotoxin in complex with Centi-3FTX-D09 antibody | | Descriptor: | ACETATE ION, Centi-3FTX-D09 Fab heavy chain, Centi-3FTX-D09 Fab light chain, ... | | Authors: | Pletnev, S, Verardi, R, Tully, E.S, Glanville, J, Kwong, P.D. | | Deposit date: | 2022-06-12 | | Release date: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Venom protection by antibody from a snakebite hyperimmune subject
To Be Published
|
|
4N3D
 
 | | Crystal structure of the dimeric variant EGFP-K162Q in P61 space group | | Descriptor: | GLYCEROL, Green fluorescent protein, PHOSPHATE ION, ... | | Authors: | Pletneva, N.V, Pletnev, V.Z, Pletnev, S.V. | | Deposit date: | 2013-10-07 | | Release date: | 2014-08-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Three dimensional structure of the dimeric gene-engineered variant of green fluorescent protein egfp-K162Q in P61 crystal space group
Rus.J.Bioorg.Chem., 40, 2014
|
|
6LIK
 
 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with Galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Iqbal, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6LI7
 
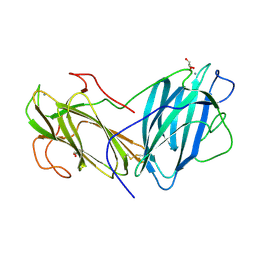 | | Crystal Structure of Lectin from Pleurotus ostreatus in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vajravijayan, S, Pletnev, S, Luo, Z, Ankur, T, Gunasekaran, K, Nandhagopal, N. | | Deposit date: | 2019-12-10 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystallographic and calorimetric analysis on Pleurotus ostreatus lectin and its sugar complexes - promiscuous binding driven by geometry.
Int.J.Biol.Macromol., 152, 2020
|
|
6ZQO
 
 | | EYFP mutant - F165G | | Descriptor: | G protein/GFP fusion protein, SULFATE ION | | Authors: | Pletnev, V.Z, Pletnev, S.V, Pletneva, N.V. | | Deposit date: | 2020-07-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Amino acid residue at the 165th position tunes EYFP chromophore maturation. A structure-based design.
Comput Struct Biotechnol J, 19, 2021
|
|
