5NL6
 
 | |
5N72
 
 | |
5NL7
 
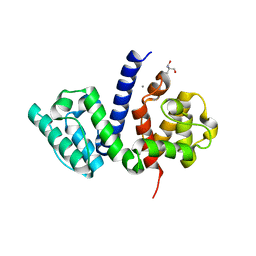 | | The crystal structure of the Actin Binding Domain (ABD) of alpha actinin from Entamoeba histolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative | | Authors: | Pinotsis, N, Djinovic-Carugo, K, Khan, M.B. | | Deposit date: | 2017-04-04 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7O1N
 
 | |
8AYS
 
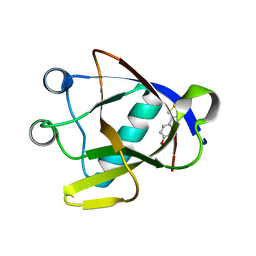 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8AZ8
 
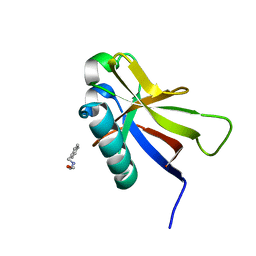 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
6HU9
 
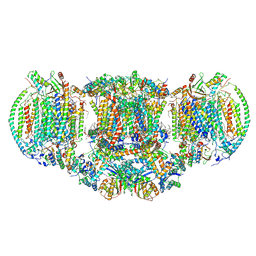 | | III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Hartley, A.M, Pinotsis, N, Marechal, A. | | Deposit date: | 2018-10-05 | | Release date: | 2018-12-26 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat. Struct. Mol. Biol., 26, 2019
|
|
2FGO
 
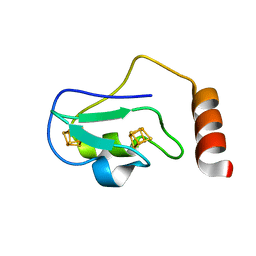 | | Structure of the 2[4FE-4S] ferredoxin from Pseudomonas aeruginosa | | Descriptor: | Ferredoxin, IRON/SULFUR CLUSTER | | Authors: | Giastas, P, Pinotsis, N, Mavridis, I.M. | | Deposit date: | 2005-12-22 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structure of the 2[4Fe-4S] ferredoxin from Pseudomonas aeruginosa at 1.32-A resolution: comparison with other high-resolution structures of ferredoxins and contributing structural features to reduction potential values.
J.Biol.Inorg.Chem., 11, 2006
|
|
3RBS
 
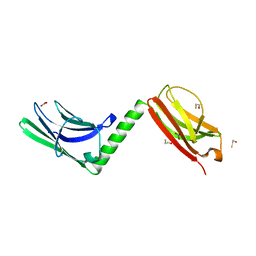 | |
8CRF
 
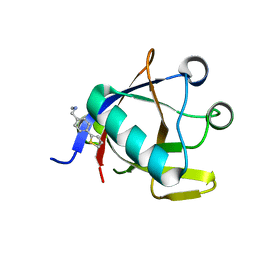 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRK
 
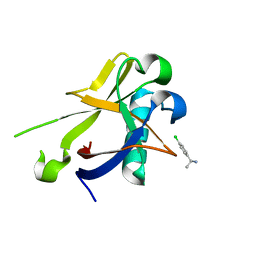 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRM
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8A55
 
 | |
7Z10
 
 | | Monomeric respiratory complex IV isolated from S. cerevisiae | | Descriptor: | COPPER (II) ION, CYTOCHROME C OXIDASE SUBUNIT 3; SYNONYM: CYTOCHROME C OXIDASE POLYPEPTIDE III, COX3, ... | | Authors: | Marechal, A, Hartley, A, Ing, G, Pinotsis, N. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Cryo-EM structure of a monomeric yeast S. cerevisiae complex IV isolated with maltosides: Implications in supercomplex formation.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7PG5
 
 | | Crystal Structure of PI3Kalpha | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.20029068 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7PG6
 
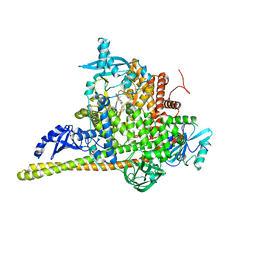 | | Crystal Structure of PI3Kalpha in complex with the inhibitor NVP-BYL719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Gong, G, Pinotsis, N, Williams, R.L, Vanhaesebroeck, B. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49943733 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
7QDF
 
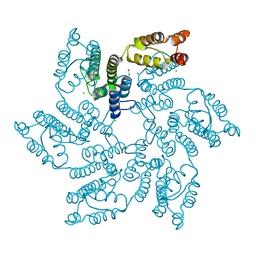 | | Hexameric HIV-1 (M-group) CA R120 mutant | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Gag polyprotein, ... | | Authors: | Govasli, M.A.L, Pinotsis, N, McAlpine-Scott, S. | | Deposit date: | 2021-11-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Evasion of cGAS and TRIM5 defines pandemic HIV.
Nat Microbiol, 7, 2022
|
|
8D3B
 
 | |
6T0B
 
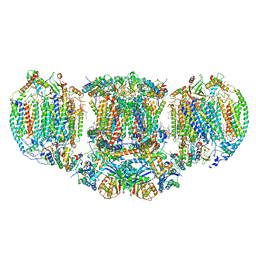 | | The III2-IV(5B)2 respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Marechal, A, Pinotsis, N, Hartley, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-22 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6T15
 
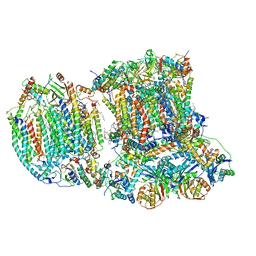 | | The III2-IV(5B)1 respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CARDIOLIPIN, COPPER (II) ION, ... | | Authors: | Marechal, A, Pinotsis, N, Hartley, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
