4R4P
 
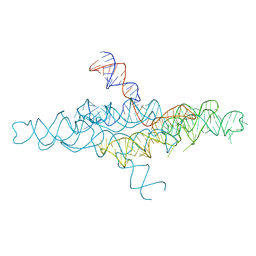 | | Crystal Structure of the VS ribozyme-A756G mutant | | 分子名称: | MAGNESIUM ION, VS ribozyme RNA | | 著者 | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
4R4V
 
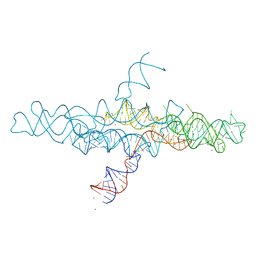 | | Crystal structure of the VS ribozyme - G638A mutant | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, VS ribozyme RNA | | 著者 | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
7U0Y
 
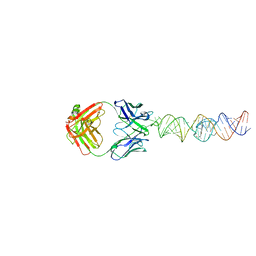 | |
8SH5
 
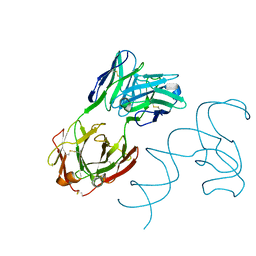 | | Crystal structure of 3'cap-independent translation enhancers (CITE) from Pea enation mosaic virus RNA 2 (PEMV2) with Fab BL3-6K170A | | 分子名称: | Fab BL3-6K170A heavy chain, Fab BL3-6K170A light chain, RNA (88-MER) | | 著者 | Lewicka, A, Roman, C, Rice, P.A, Piccirilli, J.A. | | 登録日 | 2023-04-13 | | 公開日 | 2023-08-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of a cap-independent translation enhancer RNA.
Nucleic Acids Res., 51, 2023
|
|
8UTA
 
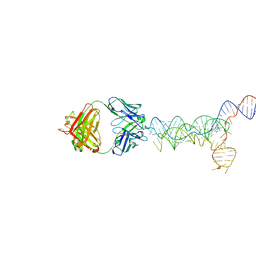 | | yjdF riboswitch from R. gauvreauii in complex with proflavine bound to Fab BL3-6 S97N | | 分子名称: | Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, MAGNESIUM ION, ... | | 著者 | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
8UIW
 
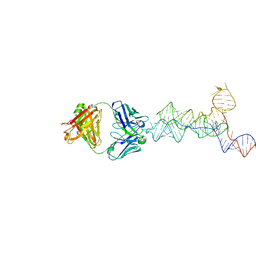 | | yjdF riboswitch from R. gauvreauii in complex with chelerythrine bound to Fab BL3-6 S97N | | 分子名称: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Fab BL3-6 S97N heavy chain, Fab BL3-6 S97N light chain, ... | | 著者 | Krochmal, D, Lewicka, A, Piccirilli, J.A. | | 登録日 | 2023-10-10 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Structural basis for promiscuity in ligand recognition by yjdF riboswitch.
Cell Discov, 10, 2024
|
|
5V3I
 
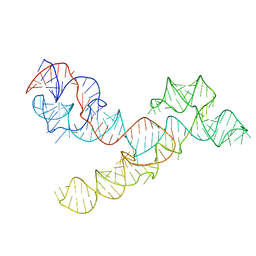 | |
5E08
 
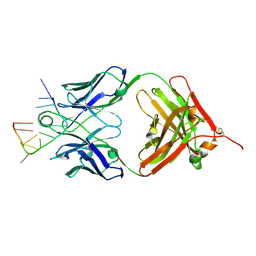 | | Specific Recognition of a Single-stranded RNA Sequence by an Engineered Synthetic Antibody Fragment | | 分子名称: | Fab Heavy Chain, Fab Light Chain, RNA | | 著者 | Huang, H, Qin, D, Li, N, Shao, Y, Staley, J.P, Kossiakoff, A.A, Koide, S, Piccirilli, J.A. | | 登録日 | 2015-09-28 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment.
J.Mol.Biol., 428, 2016
|
|
6B14
 
 | | Crystal structure of Spinach RNA aptamer in complex with Fab BL3-6S97N | | 分子名称: | Heavy chain of Fab BL3-6S97N, Light chain of Fab BL3-6S97N, MAGNESIUM ION, ... | | 著者 | DasGupta, S, Shelke, S.A, Piccirilli, J.A. | | 登録日 | 2017-09-16 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography.
Nucleic Acids Res., 46, 2018
|
|
6B3K
 
 | | Crystal structure of mutant Spinach RNA aptamer in complex with Fab BL3-6 | | 分子名称: | Heavy chain of Fab BL3-6, Light chain of Fab BL3-6, MAGNESIUM ION, ... | | 著者 | DasGupta, S, Koirala, D, Shelke, S.A, Piccirilli, J.A. | | 登録日 | 2017-09-22 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography.
Nucleic Acids Res., 46, 2018
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | 分子名称: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | 著者 | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | 登録日 | 2019-09-05 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
3OV6
 
 | | CD1c in complex with MPM (mannosyl-beta1-phosphomycoketide) | | 分子名称: | 1-O-[(S)-hydroxy{[(4S,8S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-beta-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | 著者 | Scharf, L, Li, N.S, Hawk, A.J, Garzon, D, Zhang, T, Kazen, A.R, Shah, S, Haddadian, E.J, Saghatelian, A, Faraldo-Gomez, J.D, Meredith, S.C, Piccirilli, J.A, Adams, E.J. | | 登録日 | 2010-09-15 | | 公開日 | 2011-01-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation
Immunity, 33, 2010
|
|
6DB8
 
 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | 分子名称: | 2-[(Z)-(3-methyl-1,3-benzoxazol-2(3H)-ylidene)methyl]-3-(3-sulfopropyl)-1,3-benzothiazol-3-ium, Fab-Heavy chain, Fab-Light chain, ... | | 著者 | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | 登録日 | 2018-05-02 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.86541343 Å) | | 主引用文献 | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6DB9
 
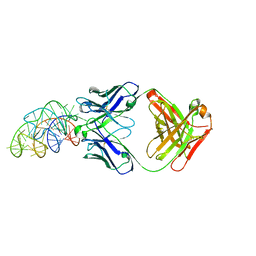 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | 分子名称: | Fab-Heavy-Chain, Fab-Light-Chain, MAGNESIUM ION, ... | | 著者 | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | 登録日 | 2018-05-02 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.025 Å) | | 主引用文献 | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6U8D
 
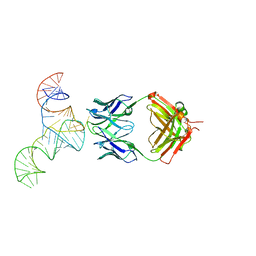 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | 分子名称: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | 著者 | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | 登録日 | 2019-09-04 | | 公開日 | 2019-12-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.807 Å) | | 主引用文献 | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
6XJQ
 
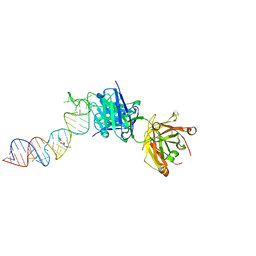 | | Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate | | 分子名称: | 2-{[(4R)-4-hydroxyhexyl]oxy}ethyl 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoate, Fab HAVx Heavy Chain, Fab HAVx Light Chain, ... | | 著者 | Koirala, D, Piccirilli, J.A. | | 登録日 | 2020-06-24 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.708 Å) | | 主引用文献 | Structural basis for substrate binding and catalysis by a self-alkylating ribozyme.
Nat.Chem.Biol., 18, 2022
|
|
6XJW
 
 | |
7SZU
 
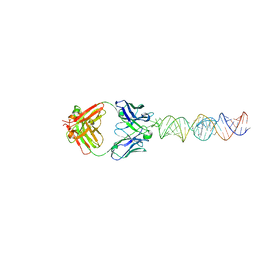 | |
6MWN
 
 | |
5C7W
 
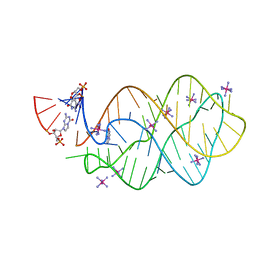 | | 5'-monophosphate Z:P Guanine Riboswitch bound to hypoxanthine. | | 分子名称: | 5'-monophosphate Z:P guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | 著者 | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | 登録日 | 2015-06-25 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.22 Å) | | 主引用文献 | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5C7U
 
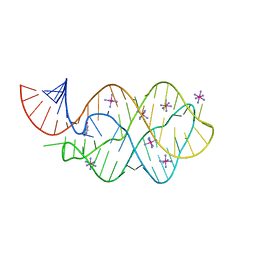 | | 5'-monophosphate wt Guanine Riboswitch bound to hypoxanthine. | | 分子名称: | 5'-monophosphate wt guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | 著者 | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | 登録日 | 2015-06-24 | | 公開日 | 2015-08-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7JRS
 
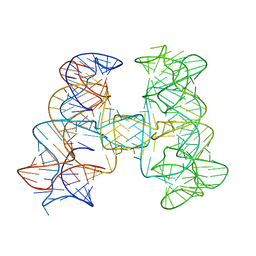 | |
7JRR
 
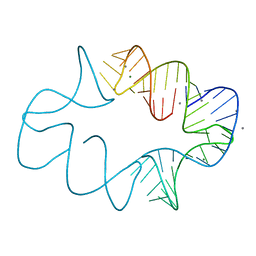 | | Crystal structures of artificially designed homomeric RNA nanoarchitectures | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, RNA (50-MER) | | 著者 | Liu, D, Shao, Y, Piccirilli, J.A, Weizmann, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structures of artificially designed discrete RNA nanoarchitectures at near-atomic resolution.
Sci Adv, 7, 2021
|
|
7JRT
 
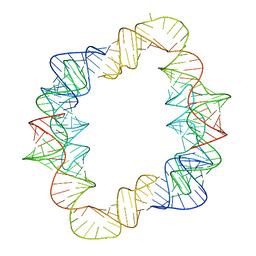 | |
6XJY
 
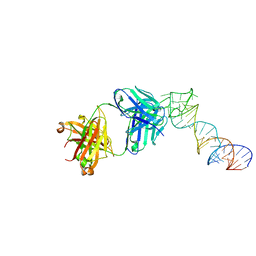 | |
