4GLQ
 
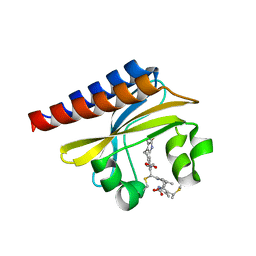 | | Crystal Structure of the blue-light absorbing form of the Thermosynechococcus elongatus PixJ GAF-domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | Authors: | Burgie, E.S, Walker, J.M, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2012-08-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | A Photo-Labile Thioether Linkage to Phycoviolobilin Provides the Foundation for the Blue/Green Photocycles in DXCF-Cyanobacteriochromes.
Structure, 21, 2013
|
|
5UHJ
 
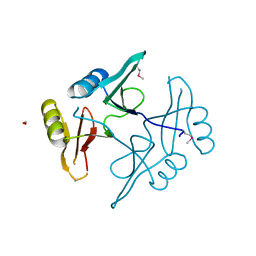 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
5UJP
 
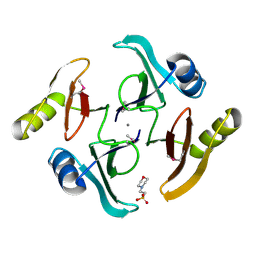 | | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The crystal structure of a glyoxalase/bleomycin resistance protein from Streptomyces sp. CB03234
To Be Published
|
|
5UMY
 
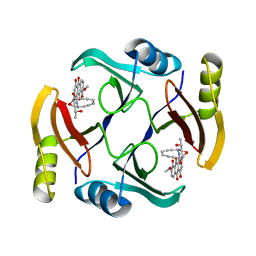 | | Crystal structure of TnmS3 in complex with tiancimycin | | Descriptor: | (1aS,11S,11aR,14Z,18R)-3,8,18-trihydroxy-11a-[(1R)-1-hydroxyethyl]-7-methoxy-11,11a-dihydro-4H-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinoline-4,9(10H)-dione, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMP
 
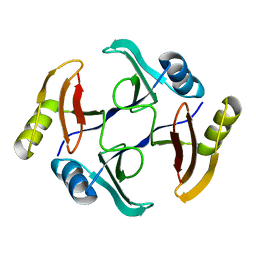 | | Crystal structure of TnmS3, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMW
 
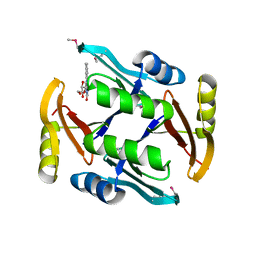 | | Crystal structure of TnmS2, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5VPJ
 
 | | The crystal structure of a thioesterase from Actinomadura verrucosospora | | Descriptor: | CHLORIDE ION, TETRAETHYLENE GLYCOL, Thioesterase | | Authors: | Tan, K, Joachimiak, G, Endres, M, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-05-05 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of a thioesterase from Actinomadura verrucosospora.
To Be Published
|
|
5UID
 
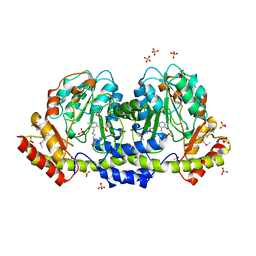 | | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus | | Descriptor: | Aminotransferase TlmJ, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Bearden, J, Phillips Jr, G.N, Joachmiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-01 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The crystal structure of an aminotransferase TlmJ from Streptoalloteichus hindustanus.
To Be Published
|
|
5UMQ
 
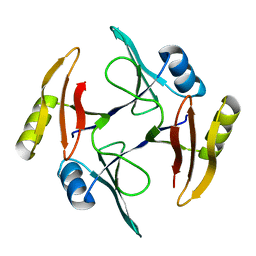 | | Crystal structure of TnmS1, an antibiotic binding protein from Streptomyces sp. CB03234 | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UMX
 
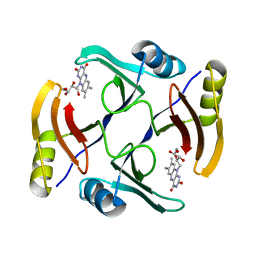 | | Crystal structure of TnmS3 in complex with riboflavin | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, RIBOFLAVIN | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-29 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Resistance to Enediyne Antitumor Antibiotics by Sequestration.
Cell Chem Biol, 25, 2018
|
|
5UNC
 
 | | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus | | Descriptor: | FORMIC ACID, L(+)-TARTARIC ACID, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus
To Be Published
|
|
2BE4
 
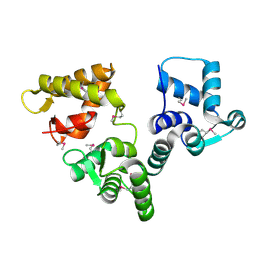 | | X-RAY STRUCTURE AN EF-HAND PROTEIN FROM DANIO RERIO Dr.36843 | | Descriptor: | hypothetical protein LOC449832 | | Authors: | Wesenberg, G.E, Phillips Jr, G.N, Han, B.W, Bitto, E, Bingman, C.A, Bae, E, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-21 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of Danio rerio secretagogin: A hexa-EF-hand calcium sensor.
Proteins, 76, 2009
|
|
5UQP
 
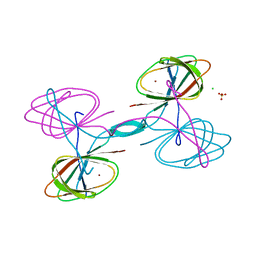 | | The crystal structure of cupin protein from Rhodococcus jostii RHA1 | | Descriptor: | CHLORIDE ION, Cupin, SULFATE ION, ... | | Authors: | Tan, K, Li, H, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of cupin protein from Rhodococcus jostii RHA1
To Be Published
|
|
4HVM
 
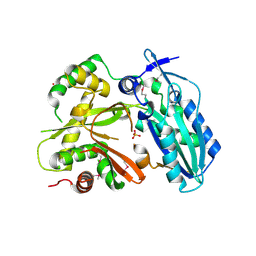 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4ISJ
 
 | | RNA Ligase RtcB in complex with Mn(II) | | Descriptor: | MANGANESE (II) ION, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.344 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4IT0
 
 | | Structure of the RNA ligase RtcB-GMP/Mn(II) complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4ISZ
 
 | | RNA ligase RtcB in complex with GTP alphaS and Mn(II) | | Descriptor: | GUANOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-01-17 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structures of the Noncanonical RNA Ligase RtcB Reveal the Mechanism of Histidine Guanylylation.
Biochemistry, 52, 2013
|
|
4K0B
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L2Z
 
 | | Crystal structure of S-Adenosylmethionine synthetase from Sulfolobus solfataricus complexed with SAE and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-05 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
1ABS
 
 | | PHOTOLYSED CARBONMONOXY-MYOGLOBIN AT 20 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Schlichting, I, Berendzen, J, Phillips Jr, G.N, Sweet, R.M. | | Deposit date: | 1997-01-28 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of photolysed carbonmonoxy-myoglobin.
Nature, 371, 1994
|
|
2Q44
 
 | | Ensemble refinement of the protein crystal structure of gene product from Arabidopsis thaliana At1g77540 | | Descriptor: | BROMIDE ION, Uncharacterized protein At1g77540 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
2Q4H
 
 | | Ensemble refinement of the crystal structure of GALT-like protein from Arabidopsis thaliana At5g18200 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Probable galactose-1-phosphate uridyl transferase, ... | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
