4CIJ
 
 | |
4CWE
 
 | |
4CWC
 
 | |
5ADU
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R, Brooke, E.J, Wehlin, S.A, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4F
 
 | | The mechanism of Hydrogen Activation by NiFe-hydrogenases. | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
5A4I
 
 | | The mechanism of Hydrogen activation by NiFE-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.C, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
4AUN
 
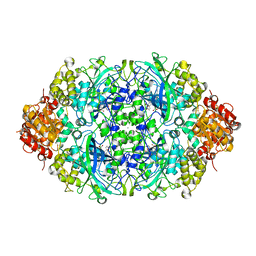 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
4AUL
 
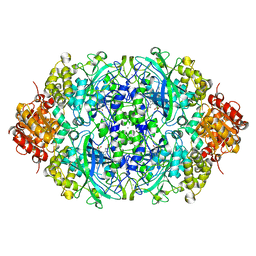 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1SPU
 
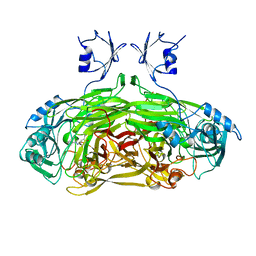 | | STRUCTURE OF OXIDOREDUCTASE | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Wilmot, C.M, Phillips, S.E.V. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction.
Biochemistry, 36, 1997
|
|
1QAK
 
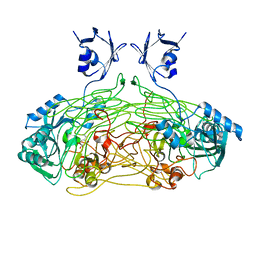 | | THE ACTIVE SITE BASE CONTROLS COFACTOR REACTIVITY IN ESCHERICHIA COLI AMINE OXIDASE : X-RAY CRYSTALLOGRAPHIC STUDIES WITH MUTATIONAL VARIANTS | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E, McPherson, M.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: x-ray crystallographic studies with mutational variants.
Biochemistry, 38, 1999
|
|
1U1Y
 
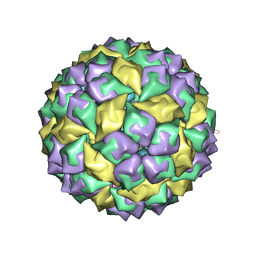 | | Crystal structure of a complex between WT bacteriophage MS2 coat protein and an F5 aptamer RNA stemloop with 2aminopurine substituted at the-10 position | | Descriptor: | 5'-R(*CP*CP*GP*GP*(2PR)P*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*G)-3', Coat protein | | Authors: | Horn, W.T, Convery, M.A, Stonehouse, N.J, Adams, C.J, Liljas, L, Phillips, S.E, Stockley, P.G. | | Deposit date: | 2004-07-16 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions.
Rna, 10, 2004
|
|
5LRY
 
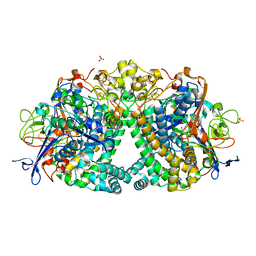 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
1JRQ
 
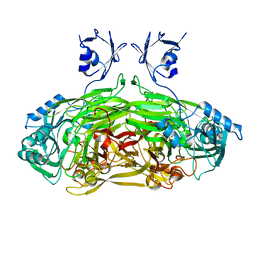 | | X-ray Structure Analysis of the Role of the Conserved Tyrosine-369 in Active Site of E. coli Amine Oxidase | | Descriptor: | CALCIUM ION, COPPER (II) ION, Copper amine oxidase | | Authors: | Murray, J.M, Kurtis, C.R, Tambarajah, W, Saysell, C.G, Wilmot, C.M, Parsons, M.R, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2001-08-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conserved tyrosine-369 in the active site of Escherichia coli copper amine oxidase is not essential.
Biochemistry, 40, 2001
|
|
1K3I
 
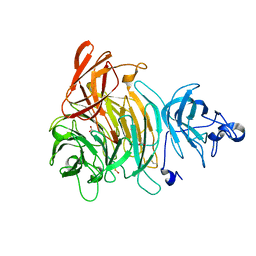 | | Crystal Structure of the Precursor of Galactose Oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, Galactose Oxidase Precursor, ... | | Authors: | Firbank, S.J, Rogers, M.S, Wilmot, C.M, Dooley, D.M, Halcrow, M.A, Knowles, P.F, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1D6Y
 
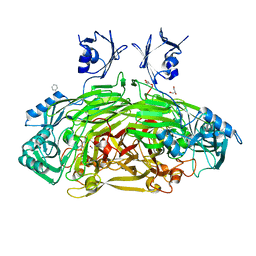 | | CRYSTAL STRUCTURE OF E. COLI COPPER-CONTAINING AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE AND COMPLEXED WITH NITRIC OXIDE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1ZNH
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTAN-1-OL | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
1ZNG
 
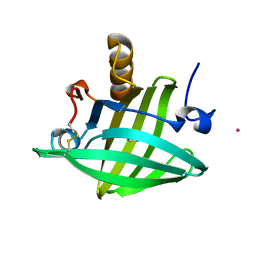 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, HEPTAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
1ZNE
 
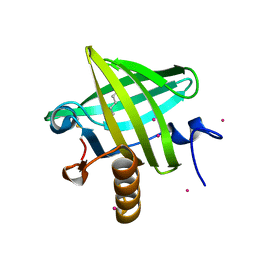 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, HEXAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
1ZNL
 
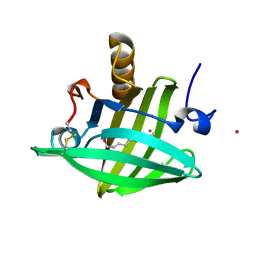 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, DECAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
1D6U
 
 | | CRYSTAL STRUCTURE OF E. COLI AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1D6Z
 
 | | CRYSTAL STRUCTURE OF THE AEROBICALLY FREEZE TRAPPED RATE-DETERMINING CATALYTIC INTERMEDIATE OF E. COLI COPPER-CONTAINING AMINE OXIDASE. | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-16 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1ZND
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, Major Urinary Protein, PENTAN-1-OL | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
1ZNK
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, Major Urinary Protein, NONAN-1-OL | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
2HWV
 
 | | Crystal structure of an essential response regulator DNA binding domain, VicRc in Enterococcus faecalis, a member of the YycF subfamily. | | Descriptor: | DNA-binding response regulator VicR, SULFATE ION | | Authors: | Trinh, C.H, Liu, Y, Phillips, S.E.V, Phillips-Jones, M.K. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the response regulator VicR DNA-binding domain.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
