2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNZ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 crystal form I | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-ETHOXYETHANOL, LACTIC ACID, ... | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNQ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in space group P21 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
1CMA
 
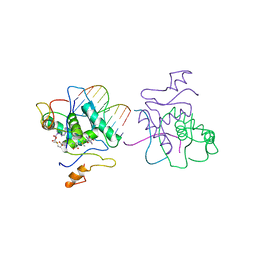 | | MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*GP*TP*CP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*AP*CP*GP*TP*CP*T)-3'), PROTEIN (MET REPRESSOR), ... | | Authors: | Somers, W.S, Phillips, S.E.V. | | Deposit date: | 1992-08-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands.
Nature, 359, 1992
|
|
2XFW
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in crystal form III | | Descriptor: | N-ACETYLNEURAMINIC ACID LYASE, PENTAETHYLENE GLYCOL, PYRUVIC ACID | | Authors: | Campeotto, I, Murshudov, G.N, Bolt, A.H, Trinh, C.H, Phillips, S.E.V, Nelson, A, Pearson, A.R, Berry, A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
To be Published
|
|
2WPB
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate and the inhibitor (2R,3R)-2,3,4- trihydroxy-N,N-dipropylbutanamide in space group P21 crystal form I | | Descriptor: | (2R,3R)-2,3,4-TRIHYDROXY-N,N-DIPROPYLBUTANAMIDE, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2YGY
 
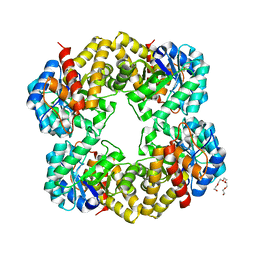 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form II | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL | | Authors: | Campeotto, I, Nelson, A, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2011-04-23 | | Release date: | 2012-04-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pathological macromolecular crystallographic data affected by twinning, partial-disorder and exhibiting multiple lattices for testing of data processing and refinement tools.
Sci Rep, 8, 2018
|
|
1K3I
 
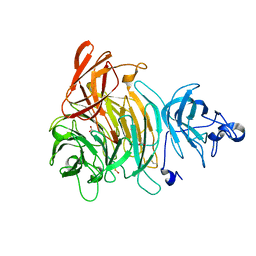 | | Crystal Structure of the Precursor of Galactose Oxidase | | Descriptor: | ACETATE ION, CALCIUM ION, Galactose Oxidase Precursor, ... | | Authors: | Firbank, S.J, Rogers, M.S, Wilmot, C.M, Dooley, D.M, Halcrow, M.A, Knowles, P.F, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the precursor of galactose oxidase: an unusual self-processing enzyme.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1MJQ
 
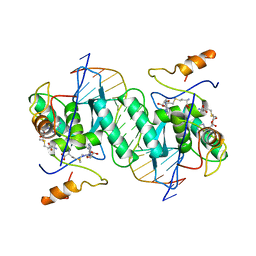 | |
1LDS
 
 | | Crystal Structure of monomeric human beta-2-microglobulin | | Descriptor: | SODIUM ION, beta-2-microglobulin | | Authors: | Trinh, C.H, Smith, D.P, Kalverda, A.P, Phillips, S.E.V, Radford, S.E. | | Deposit date: | 2002-04-09 | | Release date: | 2002-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of monomeric human beta-2-microglobulin reveals clues to its amyloidogenic properties.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MJO
 
 | |
1CMC
 
 | |
1MJP
 
 | |
1JRQ
 
 | | X-ray Structure Analysis of the Role of the Conserved Tyrosine-369 in Active Site of E. coli Amine Oxidase | | Descriptor: | CALCIUM ION, COPPER (II) ION, Copper amine oxidase | | Authors: | Murray, J.M, Kurtis, C.R, Tambarajah, W, Saysell, C.G, Wilmot, C.M, Parsons, M.R, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2001-08-14 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conserved tyrosine-369 in the active site of Escherichia coli copper amine oxidase is not essential.
Biochemistry, 40, 2001
|
|
1MJM
 
 | |
1M0D
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site and Bound Manganese Ions | | Descriptor: | Endodeoxyribonuclease I, MANGANESE (II) ION, SULFATE ION | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-12 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I.
EMBO J., 21, 2002
|
|
1EFC
 
 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1CFV
 
 | | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | | Descriptor: | ESTRONE BETA-D-GLUCURONIDE, MONOCLONAL ANTIBODY FV4155, ZINC ION | | Authors: | Trinh, C.H, Phillips, S.E.V. | | Deposit date: | 1997-04-11 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity.
Structure, 5, 1997
|
|
2LTN
 
 | | DESIGN, EXPRESSION, AND CRYSTALLIZATION OF RECOMBINANT LECTIN FROM THE GARDEN PEA (PISUM SATIVUM) | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEA LECTIN, ... | | Authors: | Suddath, F.L, Phillips, S.R, Einspahr, H. | | Deposit date: | 1990-06-26 | | Release date: | 1990-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, expression, and crystallization of recombinant lectin from the garden pea (Pisum sativum).
J.Biol.Chem., 264, 1989
|
|
1DMU
 
 | | Crystal structure of the restriction endonuclease BglI (e.c.3.1.21.4) bound to its dna recognition sequence | | Descriptor: | BETA-MERCAPTOETHANOL, BGLI RESTRICTION ENDONUCLEASE, CALCIUM ION, ... | | Authors: | Newman, M, Lunnen, K, Wilson, G, Greci, J, Schildkraut, I, Phillips, S.E.V. | | Deposit date: | 1999-12-15 | | Release date: | 1999-12-18 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of restriction endonuclease BglI bound to its interrupted DNA recognition sequence.
EMBO J., 17, 1998
|
|
1BFV
 
 | | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | | Descriptor: | ESTRIOL 3-(B-D-GLUCURONIDE), FV4155, ZINC ION | | Authors: | Trinh, C.H, Phillips, S.E.V. | | Deposit date: | 1997-05-27 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity.
Structure, 5, 1997
|
|
1M0I
 
 | | Crystal Structure of Bacteriophage T7 Endonuclease I with a Wild-Type Active Site | | Descriptor: | SULFATE ION, endodeoxyribonuclease I | | Authors: | Hadden, J.M, Declais, A.C, Phillips, S.E, Lilley, D.M. | | Deposit date: | 2002-06-13 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal ions bound at the active site of the junction-resolving enzyme T7 endonuclease I
Embo J., 21, 2002
|
|
1AUA
 
 | | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHATIDYLINOSITOL TRANSFER PROTEIN SEC14P, octyl beta-D-glucopyranoside | | Authors: | Sha, B, Phillips, S.E, Bankaitis, V.A, Luo, M. | | Deposit date: | 1997-08-20 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein.
Nature, 391, 1998
|
|
1DYU
 
 | | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2000-02-08 | | Release date: | 2000-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Active Site Base Controls Cofactor Reactivity in Escherichia Coli Amine Oxidase : X-Ray Crystallographicstudies with Mutational Variants
Biochemistry, 38, 1999
|
|
