3FWY
 
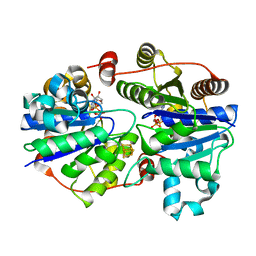 | | Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase (BchL) with MgADP bound: a homologue of the nitrogenase Fe protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein, ... | | Authors: | Sarma, R, Barney, B.M, Hamilton, T.L, Jones, A, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the L Protein of Rhodobacter sphaeroides Light-Independent Protochlorophyllide Reductase with MgADP Bound: A Homologue of the Nitrogenase Fe Protein.
Biochemistry, 47, 2008
|
|
5JFC
 
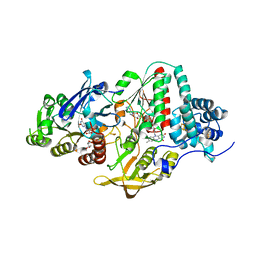 | | NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, King, P.W, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
5JCA
 
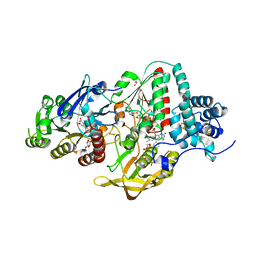 | | NADP(H) bound NADH-dependent Ferredoxin:NADP Oxidoreductase (NfnI) from Pyrococcus furiosus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Zadvornyy, O.A, Schut, G.J, Nguyen, D.M, Artz, J.H, Tokmina-Lukaszewska, M, Lipscomb, G, Adams, M.W, Peters, J.W. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into energy conservation by flavin-based electron bifurcation.
Nat. Chem. Biol., 13, 2017
|
|
4UN2
 
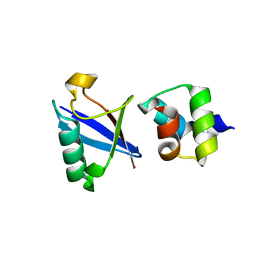 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2M6N
 
 | | 3D solution structure of EMI1 (Early Mitotic Inhibitor 1) | | Descriptor: | F-box only protein 5, ZINC ION | | Authors: | Frye, J.J, Brown, N.G, Petzold, G, Watson, E.R, Royappa, G.R, Nourse, A, Jarvis, M, Kriwacki, R.W, Peters, J, Stark, H, Schulman, B.A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MT5
 
 | | Isolated Ring domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissman, F, Royappa, G, Schulman, B, Jarvis, M, Vanderlinden, R, Frye, J.J, Qiao, R, Petzold, G, Peters, J, Stark, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
4Z1Y
 
 | |
4Z17
 
 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
4YWS
 
 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
6N6P
 
 | | Crystal structure of [FeFe]-hydrogenase in the presence of 7 mM Sodium dithionite | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Peters, J.W. | | Deposit date: | 2018-11-26 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
4R2Y
 
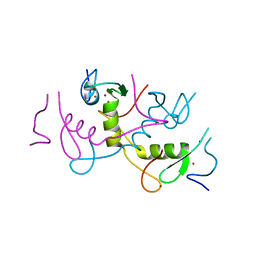 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
6NAC
 
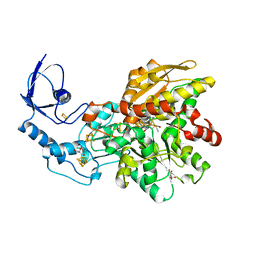 | | Crystal structure of [FeFe]-hydrogenase I (CpI) solved with single pulse free electron laser data | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Cohen, A.E, Davidson, C.M, Zadvornyy, O.A, Keable, S.M, Lyubimov, A.Y, Song, J, McPhillips, S.E, Soltis, S.M, Peters, J.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6N59
 
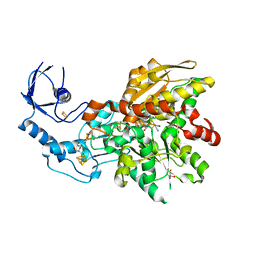 | | 1.0 Angstrom crystal structure of [FeFe]-hydrogenase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Artz, J.H, Peters, J.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Tuning Catalytic Bias of Hydrogen Gas Producing Hydrogenases.
J.Am.Chem.Soc., 142, 2020
|
|
6DO0
 
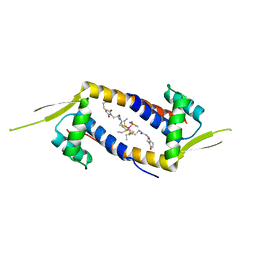 | | Crystal structure of the multidrug binding transcriptional regulator LmrR in complex with Rhodium Bis-diphosphine Complex | | Descriptor: | Transcriptional regulator, PadR-like family, bis[diethyl(methyl)-lambda~5~-phosphanyl]{bis[{[(2-{[2-(2,5-dioxopyrrolidin-1-yl)ethyl]amino}-2-oxoethyl)amino]methyl}(diethyl)-lambda~5~-phosphanyl]}rhodium | | Authors: | Zadvornyy, O.A, Laureanti, J.A, Katipamula, S, O'Hagan, M, Peters, J.W. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Protein Scaffold Activates Catalytic CO2 Hydrogenation by a Rhodium Bis(diphosphine) Complex
Acs Catalysis, 9, 2019
|
|
6CDK
 
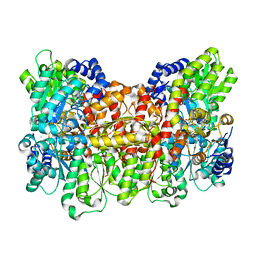 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
4R0V
 
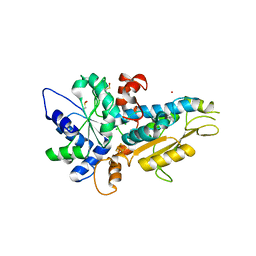 | | [FeFe]-hydrogenase Oxygen Inactivation is Initiated by the Modification and Degradation of the H cluster 2Fe Subcluster | | Descriptor: | ARSENIC, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Swanson, S.D, Ratzloff, M.W, Mulder, D.W, Artz, J.H, Ghose, S, Hoffman, A, White, S, Zadvornyy, O.A, Broderick, J.B, Bothner, B, King, P.W, Peters, J.W. | | Deposit date: | 2014-08-01 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | [FeFe]-Hydrogenase Oxygen Inactivation Is Initiated at the H Cluster 2Fe Subcluster.
J.Am.Chem.Soc., 137, 2015
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6CHR
 
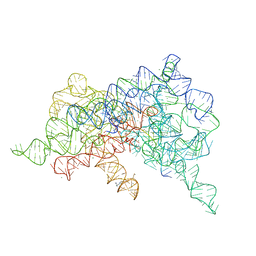 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | Descriptor: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | Authors: | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
6BBL
 
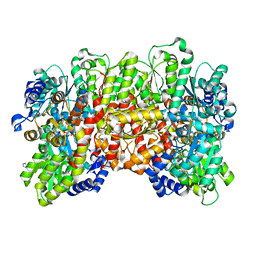 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
3PND
 
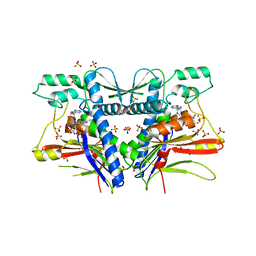 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
3LX4
 
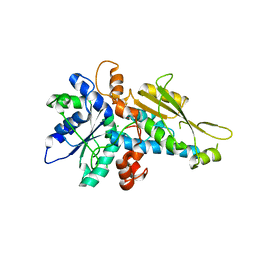 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
5VJ7
 
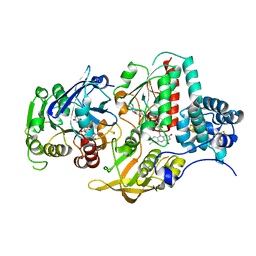 | | Ferredoxin NADP Oxidoreductase (Xfn) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP(+) reductase subunit alpha, ... | | Authors: | Zadvornyy, O.A, Nguyen, D.M.N, Schut, G.J, Lipscomb, G.L, Tokmina-Lukaszewska, M, Adams, M.W.W, Peters, J.W. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two functionally distinct NADP(+)-dependent ferredoxin oxidoreductases maintain the primary redox balance of Pyrococcus furiosus.
J. Biol. Chem., 292, 2017
|
|
5L9U
 
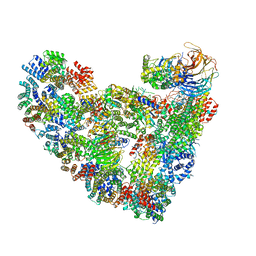 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
