1HQP
 
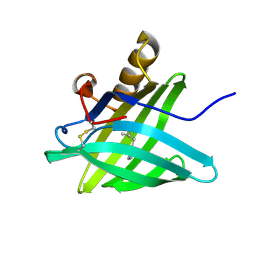 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF PORCINE ODORANT-BINDING PROTEIN | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, ODORANT-BINDING PROTEIN | | Authors: | Perduca, M, Mancia, F, Del Giorgio, R, Monaco, H.L. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a truncated form of porcine odorant-binding protein.
Proteins, 42, 2001
|
|
9GNS
 
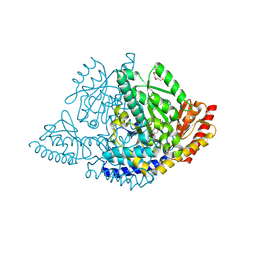 | |
9HRH
 
 | |
9HRI
 
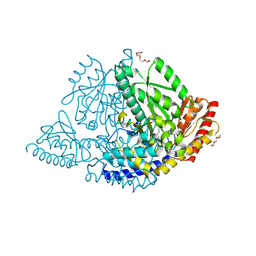 | |
6STR
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 60 seconds soaking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STQ
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 30 seconds soaking | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STN
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STM
 
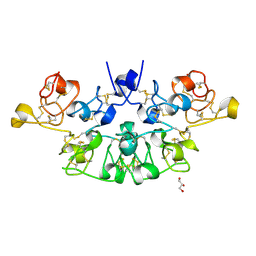 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STP
 
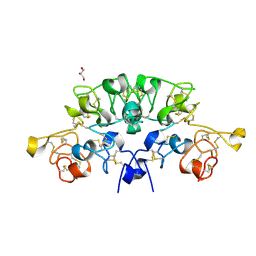 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with sialic acid | | Descriptor: | Arundo donax Lectin (ADL), GLYCEROL, N-acetyl-alpha-neuraminic acid | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STO
 
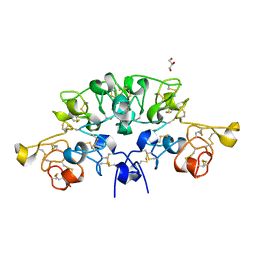 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL, ... | | Authors: | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | Deposit date: | 2019-09-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
5NU9
 
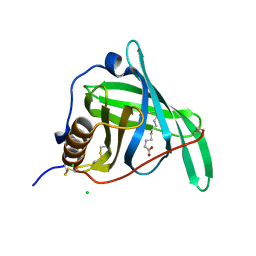 | |
5NU8
 
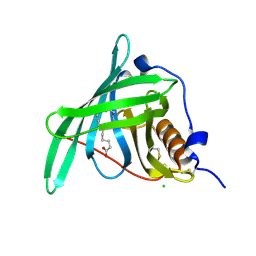 | |
5NUB
 
 | |
5NU7
 
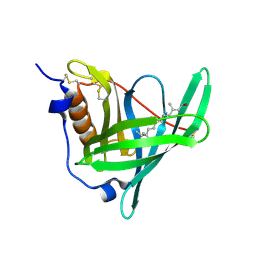 | | Structure of human holo plasma RBP4 | | Descriptor: | CHLORIDE ION, RETINOL, Retinol-binding protein 4 | | Authors: | Perduca, M, Monaco, H.L, Galliano, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human plasma retinol-binding protein (RBP4) is also a fatty acid-binding protein.
Biochim. Biophys. Acta, 1863, 2018
|
|
5NU2
 
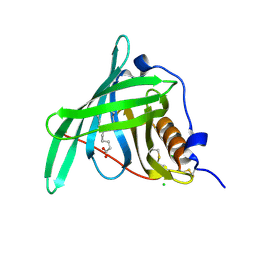 | |
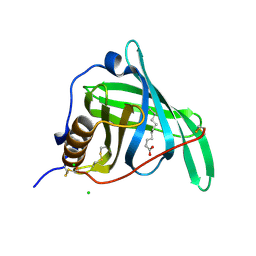
5NUA
 
 | |
5NTY
 
 | |
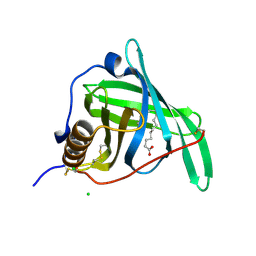
5NU6
 
 | |
4OS3
 
 | | Three-dimensional structure of the C65A-W112F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ORW
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS8
 
 | | Three-dimensional structure of the C65A-W54F-W112F triple mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ORX
 
 | | Three-dimensional structure of the C65A-K59A double mutant of Human lipocalin-type Prostaglandin D Synthase holo-form | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ORR
 
 | | Threedimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase olo-form | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OS0
 
 | | Three-dimensional structure of the C65A-W54F double mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ORS
 
 | | Three-dimensional structure of the C65A mutant of Human lipocalin-type Prostaglandin D Synthase apo-form | | Descriptor: | Prostaglandin-H2 D-isomerase, SULFATE ION | | Authors: | Perduca, M, Bovi, M, Bertinelli, M, Bertini, E, Destefanis, L, Carrizo, M.E, Capaldi, S, Monaco, H.L. | | Deposit date: | 2014-02-12 | | Release date: | 2014-08-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structures of mutants of residues that affect access to the ligand-binding cavity of human lipocalin-type prostaglandin D synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
