4OZT
 
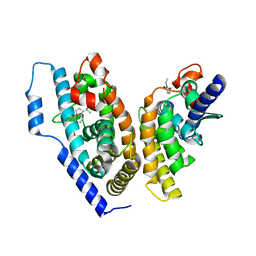 | | crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (PonA crystal) | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, N-ETHYLMALEIMIDE, ... | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5KR4
 
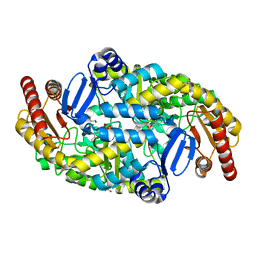 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR6
 
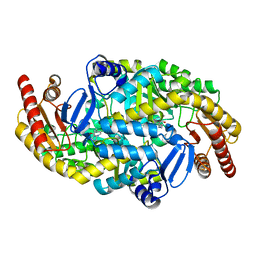 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQU
 
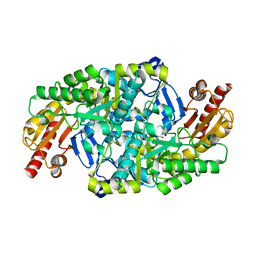 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQT
 
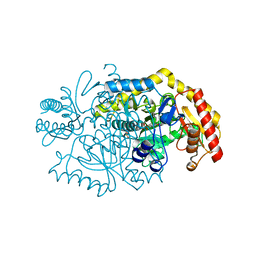 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminoburyrate transaminase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR3
 
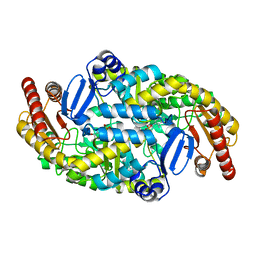 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQW
 
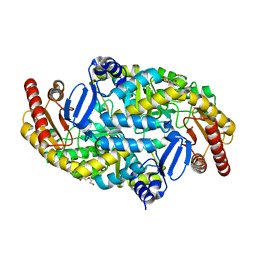 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR5
 
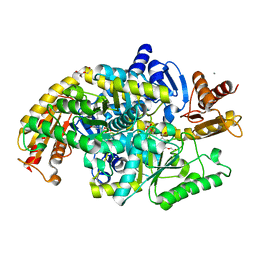 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | Descriptor: | 4-aminobutyrate transaminase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
1BN6
 
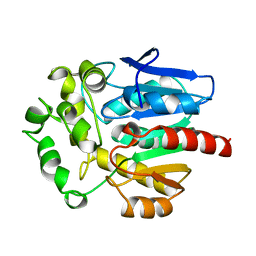 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
1BN7
 
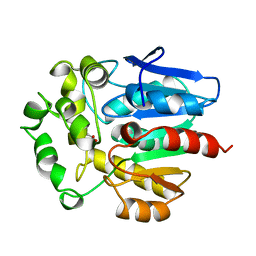 | | HALOALKANE DEHALOGENASE FROM A RHODOCOCCUS SPECIES | | Descriptor: | ACETATE ION, HALOALKANE DEHALOGENASE | | Authors: | Newman, J, Peat, T.S, Richard, R, Kan, L, Swanson, P.E, Affholter, J.A, Holmes, I.H, Schindler, J.F, Unkefer, C.J, Terwilliger, T.C. | | Deposit date: | 1998-07-31 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Haloalkane dehalogenases: structure of a Rhodococcus enzyme.
Biochemistry, 38, 1999
|
|
6BA2
 
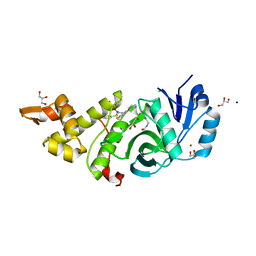 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor | | Descriptor: | 4-fluoro-5-methyl-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hermans, S.J, Chung, M.C, Peat, T.S, Baell, J.B, Thomas, T, Parker, M.W. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85003817 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
6BA4
 
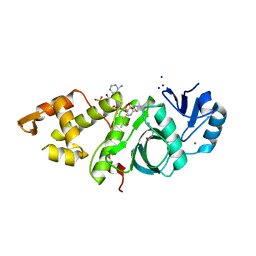 | | Crystal structure of MYST acetyltransferase domain in complex with Acetyl-CoA cofactor | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase KAT8, S-{(3S,5R,9R)-1-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} ethanethioate, ... | | Authors: | Hermans, S.J, Chung, M.C, Peat, T.S, Baell, J.B, Thomas, T, Parker, M.W. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
6CT2
 
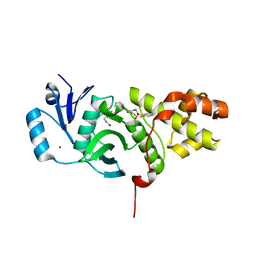 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
7M23
 
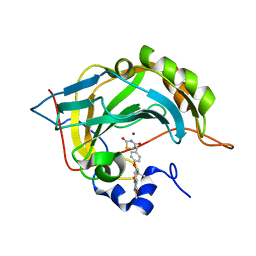 | | Human carbonic anhydrase II in complex with troglitazone | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
7M26
 
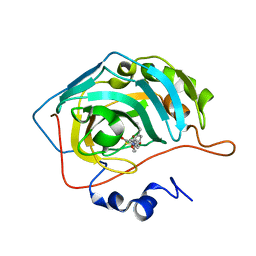 | | Human carbonic anhydrase II in complex with pioglitazone | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
7M24
 
 | |
6E3Y
 
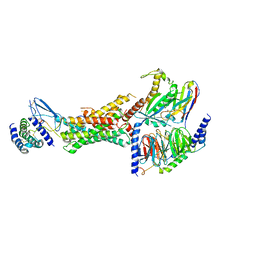 | | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor | | Descriptor: | Calcitonin gene-related peptide 1, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Deganutti, G, Glukhova, A, Koole, C, Peat, T.S, Radjainia, M, Plitzko, J.M, Baumeister, W, Miller, L.J, Hay, D.L, Christopoulos, A, Reynolds, C.A, Wootten, D, Sexton, P.M. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor.
Nature, 561, 2018
|
|
5ETK
 
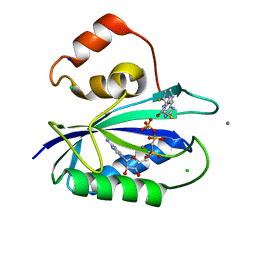 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.09 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(2-fluorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETO
 
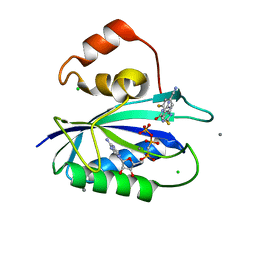 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.07 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-[(2-azanyl-6-oxidanylidene-1,9-dihydropurin-8-yl)sulfanylmethyl]-3-fluoranyl-benzenecarbonitrile, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETL
 
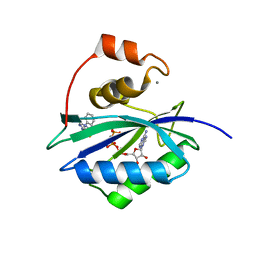 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.82 angstrom resolution | | Descriptor: | 2-[(2-azanyl-6-oxidanylidene-3,9-dihydropurin-8-yl)sulfanylmethyl]benzenecarbonitrile, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETV
 
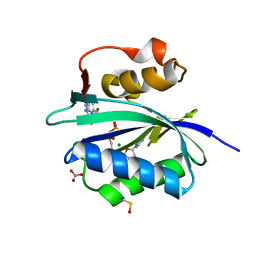 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.72 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[2-(4-bromophenyl)-2-oxidanylidene-ethyl]sulfanyl-1,9-dihydropurin-6-one, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETS
 
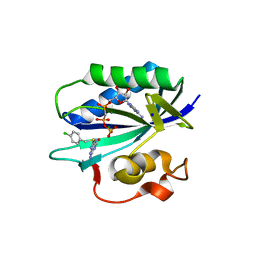 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.95 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[(4-chlorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETT
 
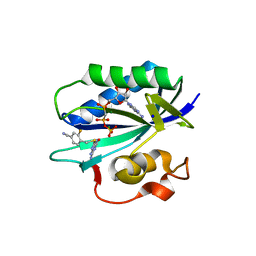 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.55 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-[(2-azanyl-6-oxidanylidene-1,9-dihydropurin-8-yl)sulfanylmethyl]-3-fluoranyl-benzenecarbonitrile, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETQ
 
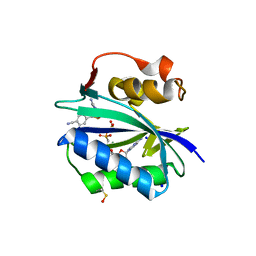 | | S. aureus 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.96 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 4-{[(2-amino-6-oxo-6,9-dihydro-1H-purin-8-yl)sulfanyl]methyl}benzonitrile, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
5ETP
 
 | | E. coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase complexed with AMPCPP and inhibitor at 1.05 angstrom resolution | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 2-azanyl-8-[2-(4-bromophenyl)-2-oxidanylidene-ethyl]sulfanyl-1,9-dihydropurin-6-one, CALCIUM ION, ... | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Basis for the Selective Binding of Inhibitors to 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase from Staphylococcus aureus and Escherichia coli.
J.Med.Chem., 59, 2016
|
|
