1H0C
 
 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
2BYE
 
 | | NMR solution structure of phospholipase c epsilon RA 1 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2BYF
 
 | | NMR solution structure of phospholipase c epsilon RA 2 domain | | Descriptor: | PHOSPHOLIPASE C, EPSILON 1 | | Authors: | Bunney, T.D, Harris, R, Gandarillas, N.L, Josephs, M.B, Roe, S.M, Paterson, H.F, Rodrigues-Lima, F, Esposito, D, Gieschik, P, Pearl, L.H, Driscoll, P.C, Katan, M. | | Deposit date: | 2005-08-01 | | Release date: | 2006-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon.
Mol.Cell, 21, 2006
|
|
2C5L
 
 | | Structure of PLC epsilon Ras association domain with hRas | | Descriptor: | GLYCEROL, GTPASE HRAS, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roe, S.M, Bunney, T.D, Katan, M, Pearl, L.H. | | Deposit date: | 2005-10-27 | | Release date: | 2006-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insights Into Ras Association Domains of Phospholipase C Epsilon
Mol.Cell, 21, 2006
|
|
2BRF
 
 | |
2BRC
 
 | | Structure of a Hsp90 Inhibitor bound to the N-terminus of Yeast Hsp90. | | Descriptor: | 4-[4-(2,3-DIHYDRO-1,4-BENZODIOXIN-6-YL)-3-METHYL-1H-PYRAZOL-5-YL]-6-ETHYLBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
2BRE
 
 | | STRUCTURE OF A HSP90 INHIBITOR BOUND TO THE N-TERMINUS OF YEAST HSP90. | | Descriptor: | 4-{4-[4-(3-AMINOPROPOXY)PHENYL]-1H-PYRAZOL-5-YL}-6-CHLOROBENZENE-1,3-DIOL, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Pearl, L.H, Prodromou, C. | | Deposit date: | 2005-05-04 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The identification, synthesis, protein crystal structure and in vitro biochemical evaluation of a new 3,4-diarylpyrazole class of Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 15, 2005
|
|
2C2V
 
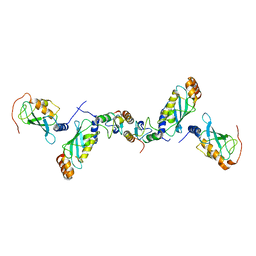 | | Crystal structure of the CHIP-UBC13-UEV1a complex | | Descriptor: | STIP1 homology and U box-containing protein 1, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex.
Mol. Cell, 20, 2005
|
|
2CGE
 
 | |
2CN8
 
 | |
2C2L
 
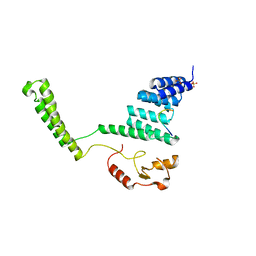 | | Crystal structure of the CHIP U-box E3 ubiquitin ligase | | Descriptor: | CARBOXY TERMINUS OF HSP70-INTERACTING PROTEIN, HSP90, NICKEL (II) ION, ... | | Authors: | Zhang, M, Roe, S.M, Pearl, L.H. | | Deposit date: | 2005-09-29 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chaperoned Ubiquitylation-Crystal Structures of the Chip U Box E3 Ubiquitin Ligase and a Chip-Ubc13-Uev1A Complex
Mol.Cell, 20, 2005
|
|
2CG9
 
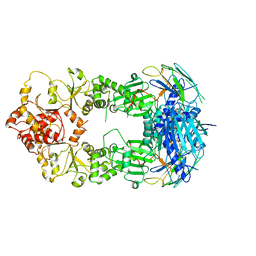 | | Crystal structure of an Hsp90-Sba1 closed chaperone complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, CO-CHAPERONE PROTEIN SBA1 | | Authors: | Ali, M.M.U, Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of an Hsp90-Nucleotide-P23/Sba1 Closed Chaperone Complex
Nature, 440, 2006
|
|
2CN5
 
 | |
2CGF
 
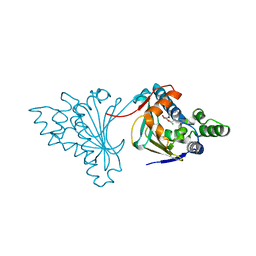 | | A RADICICOL ANALOGUE BOUND TO THE ATP BINDING SITE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-3,4,7,8,9,10-HEXAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-1,11(12H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2006-03-02 | | Release date: | 2006-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
1OE6
 
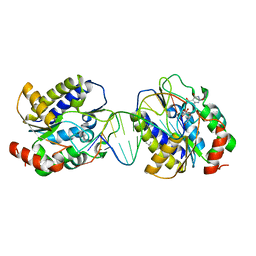 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*G)-3', 5-HYDROXYMETHYL URACIL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1OE5
 
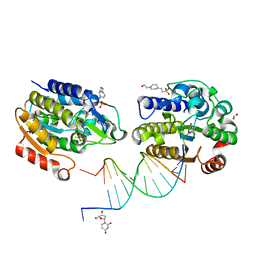 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 2'-DEOXYURIDINE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-D(*CP*3DRP*GP*GP*AP*CP*TP*3DRP*AP*CP*GP*GP*GP)-3', ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1MUG
 
 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | Descriptor: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 1998-07-10 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1ODG
 
 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | Descriptor: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | Authors: | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | Deposit date: | 2003-02-19 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
1O9U
 
 | |
1Q2Z
 
 | | The 3D solution structure of the C-terminal region of Ku86 | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Harris, R, Esposito, D, Sankar, A, Maman, J.D, Hinks, J.A, Pearl, L.H, Driscoll, P.C. | | Deposit date: | 2003-07-28 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The 3D Solution Structure of the C-terminal Region of Ku86 (Ku86CTR)
J.Mol.Biol., 335, 2004
|
|
1MWI
 
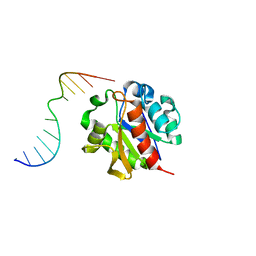 | | Crystal structure of a MUG-DNA product complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1OE4
 
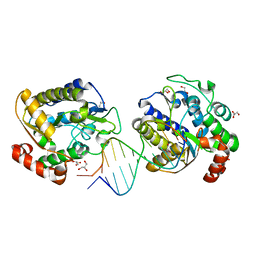 | | Xenopus SMUG1, an anti-mutator uracil-DNA Glycosylase | | Descriptor: | 5'-D(*CP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*G)-3', 5'-D(*CP*GP*GP*AP*CP*TP*3DR*AP*CP*GP*GP*G)-3', GLYCEROL, ... | | Authors: | Wibley, J.E.A, Pearl, L.H. | | Deposit date: | 2003-03-19 | | Release date: | 2003-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Specificity of the Vertebrate Anti-Mutator Uracil-DNA Glycosylase Smug1
Mol.Cell, 11, 2003
|
|
1MWJ
 
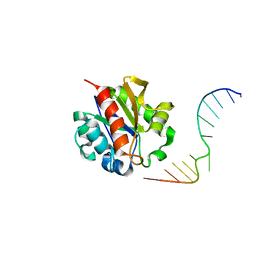 | | Crystal Structure of a MUG-DNA pseudo substrate complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*A*GP*(DU)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Scharer, O, Savva, R, Brown, T, Jiricny, J, Verdine, G.L, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of a thwarted mismatch glycosylase DNA repair complex
Embo J., 18, 1999
|
|
1MTL
 
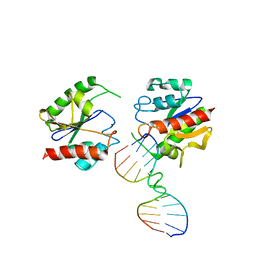 | | Non-productive MUG-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Barlow, T, Brown, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-21 | | Release date: | 2002-09-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Nat.Struct.Biol., 5, 1998
|
|
4BB7
 
 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
